Generating background points
In this vignette, we will generate some background points (pseudo-absences) using the different algorithms present in the package. The pseudo-absences are generated through the PseudoAbsences component package.
using SpeciesDistributionToolkit
using CairoMakieIn order to work on a region that is not too big, we will define our spatial extent:
spatial_extent = (left = 8.412, bottom = 41.325, right = 9.662, top = 43.060)(left = 8.412, bottom = 41.325, right = 9.662, top = 43.06)Pseudo-absence generation requires occurrences super-imposed on a layer, so we will collect a few occurrences:
species = taxon("Sitta whiteheadi"; strict = false)
query = [
"occurrenceStatus" => "PRESENT",
"hasCoordinate" => true,
"decimalLatitude" => (spatial_extent.bottom, spatial_extent.top),
"decimalLongitude" => (spatial_extent.left, spatial_extent.right),
"limit" => 300,
]
presences = occurrences(species, query...)
for i in 1:3
occurrences!(presences)
endWe will get a single layer (temperature) from CHELSA1.
dataprovider = RasterData(CHELSA1, BioClim)
temperature = 0.1SDMLayer(dataprovider; layer = "BIO1", spatial_extent...)🗺️ A 209 × 151 layer with 14432 Float64 cells
Projection: +proj=longlat +datum=WGS84 +no_defsPseudo-absences generations always starts by masking a layer by the observations. The output of this command is a layer with Boolean values, where the cells in which at least one occurrence is reported are true.
presencelayer = mask(temperature, presences)🗺️ A 209 × 151 layer with 14432 Bool cells
Projection: +proj=longlat +datum=WGS84 +no_defsWe can for example generate a buffer for pseudo-absences in a radius of 30km around each point. Note that the WithinRadius method uses kilometers and not minutes of arc, so that the actual area is the same regardless of the latitude of the points. Note that the speed of the operation depends on the number of cells with an observation (linearly), and of the radius and raster resolution (to a power of 2). Internally, the code uses a variety of tricks to only look at cells that are susceptible to being pseudo-absences, but the WithinRadius method in particular can take a bit of time.
background = pseudoabsencemask(WithinRadius, presencelayer; distance = 30.0)🗺️ A 209 × 151 layer with 14432 Bool cells
Projection: +proj=longlat +datum=WGS84 +no_defsThe pseudo-absence generation functions will return a mask, i.e. a boolean layer where the cells in which we can place a pseudo-absence are true, and the rest of the cells are false. This is useful for a variety of reasons, including adding more and more constraints to the locations of pseudo-absences. For example, we can decide that we do not want background points too close to the actual observations, and put a buffer around each.
buffer = pseudoabsencemask(WithinRadius, presencelayer; distance = 5.0)🗺️ A 209 × 151 layer with 14432 Bool cells
Projection: +proj=longlat +datum=WGS84 +no_defsWe can now exclude the data that are in the buffer:
bgmask = (!buffer) & background🗺️ A 209 × 151 layer with 14432 Bool cells
Projection: +proj=longlat +datum=WGS84 +no_defsFinally, we can plot the area in which we can put pseudo-absences as a shaded region over the layer, and plot all known occurrences as well:
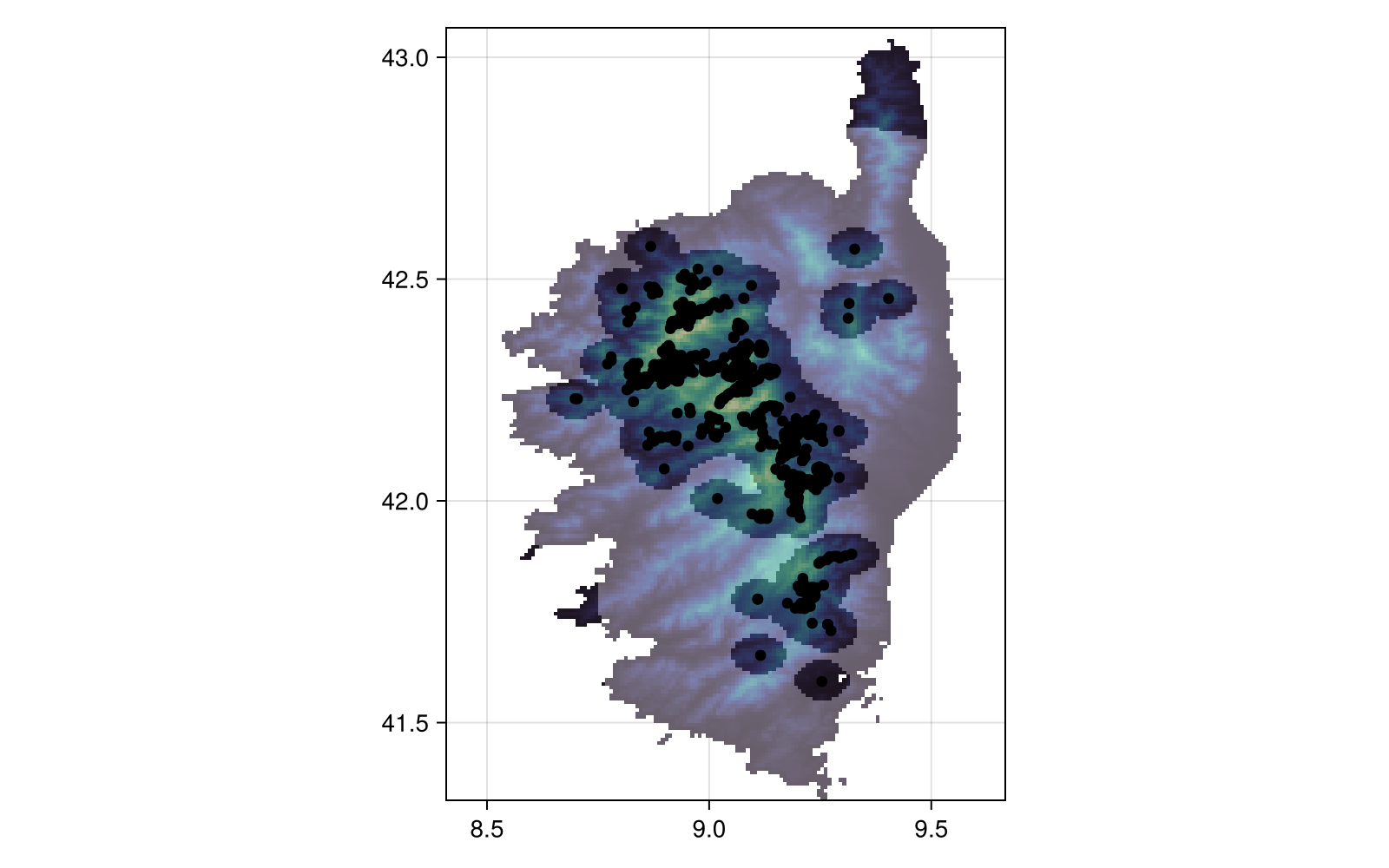
Code for the figure
heatmap(
temperature;
colormap = :deep,
axis = (; aspect = DataAspect()),
figure = (; size = (800, 500)),
)
heatmap!(bgmask; colormap = cgrad([:transparent, :white]; alpha = 0.3))
scatter!(presences; color = :black)There are additional ways to produce pseudo-absences mask, notably the surface range envelope method, which uses the bounding box of observations to allow pseudo-absences:
sre = pseudoabsencemask(SurfaceRangeEnvelope, presencelayer)🗺️ A 209 × 151 layer with 14432 Bool cells
Projection: +proj=longlat +datum=WGS84 +no_defs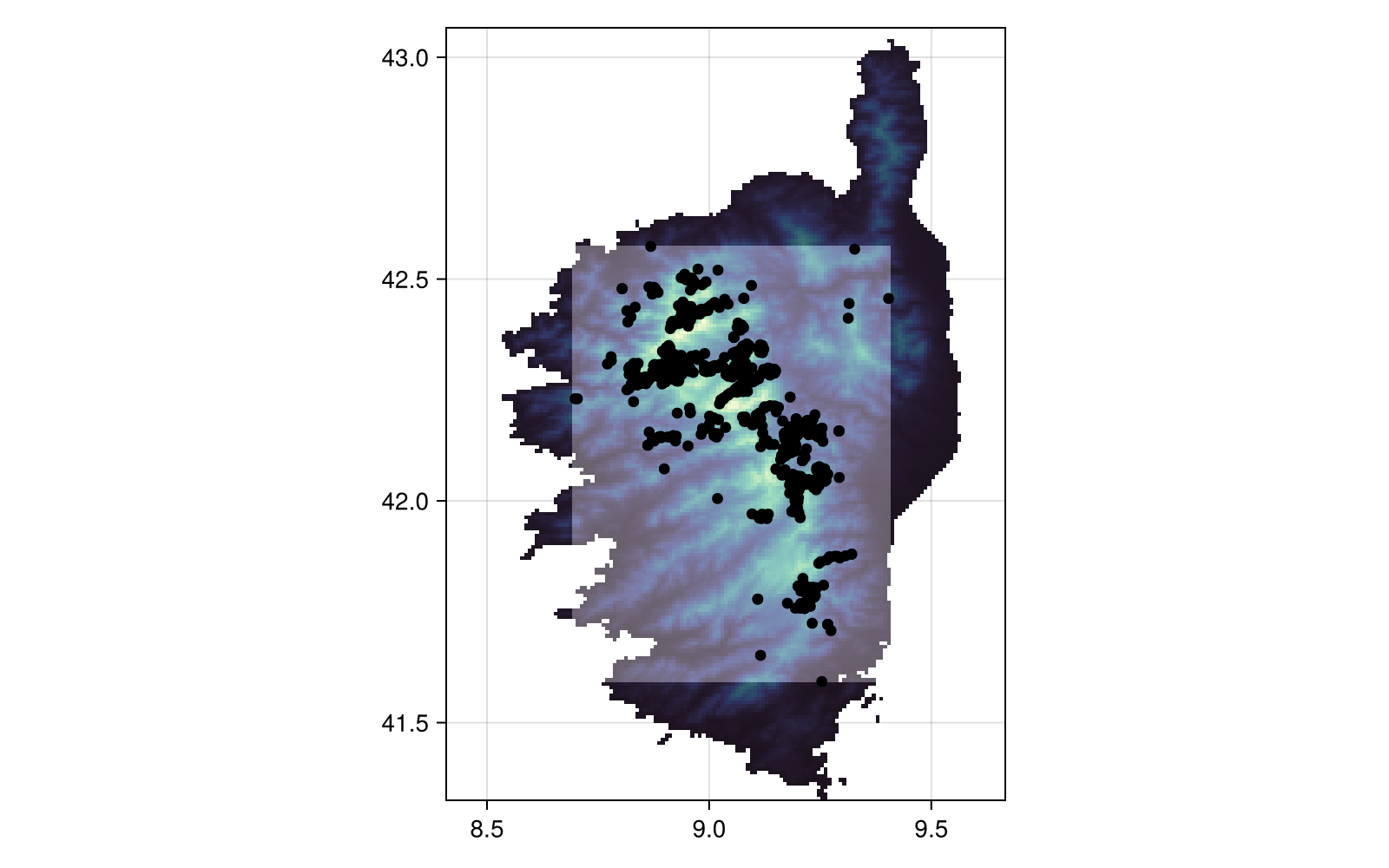
Code for the figure
heatmap(
temperature;
colormap = :deep,
axis = (; aspect = DataAspect()),
figure = (; size = (800, 500)),
)
heatmap!(sre; colormap = cgrad([:transparent, :white]; alpha = 0.3))
scatter!(presences; color = :black)The RandomSelection method (not shown) uses the entire surface of the layer as a possible pseudo-absence location.
Note that we are not yet generating pseudo-absences, and in order to do so, we need to sample the mask generated by pseudoabsencemask. We can do so using backgroundpoints, which uses the StatsBase.sample function internally.
bgpoints = backgroundpoints(bgmask, sum(presencelayer))🗺️ A 209 × 151 layer with 14432 Bool cells
Projection: +proj=longlat +datum=WGS84 +no_defsAnd finally, we can make a plot:
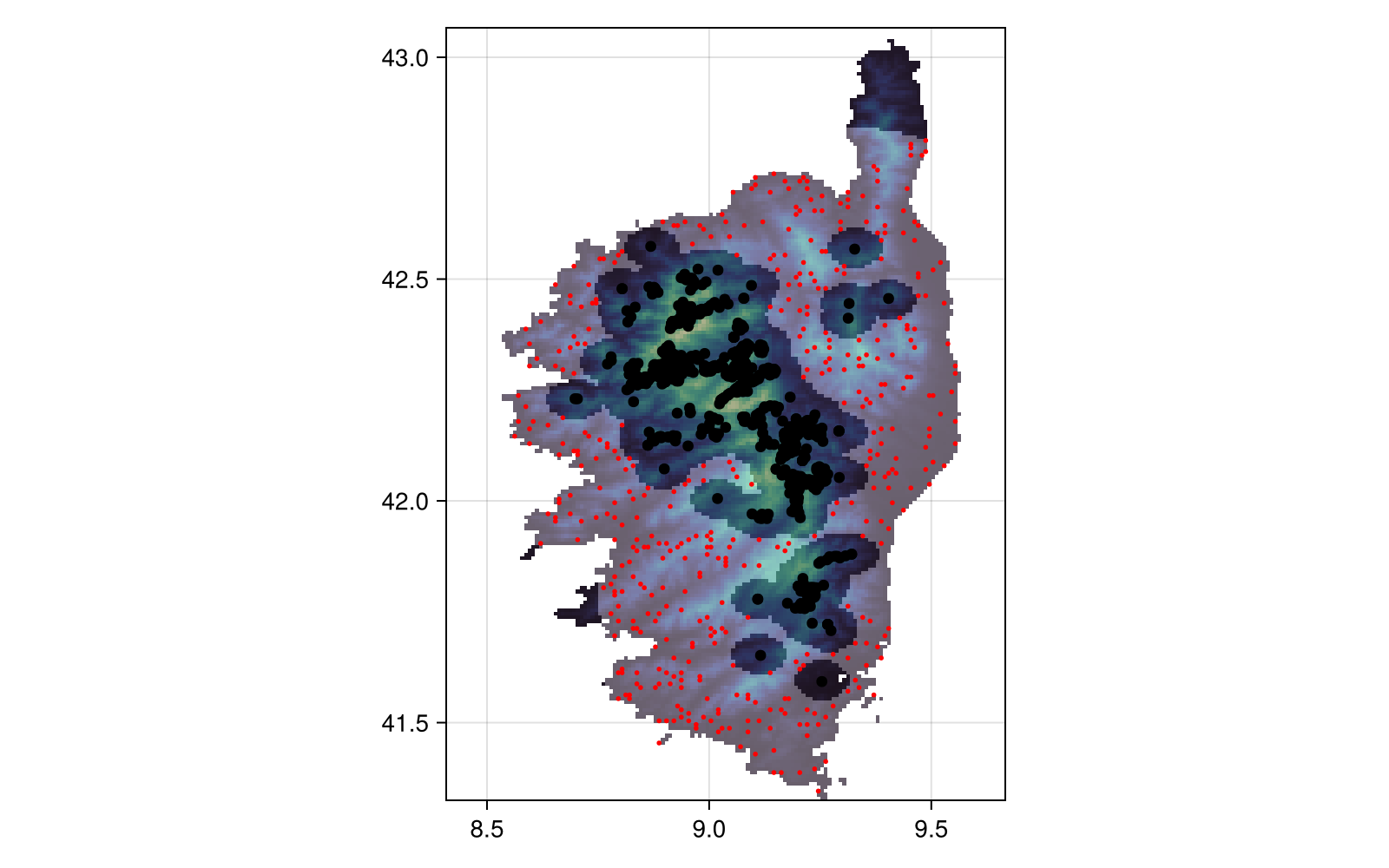
Code for the figure
heatmap(
temperature;
colormap = :deep,
axis = (; aspect = DataAspect()),
figure = (; size = (800, 500)),
)
heatmap!(bgmask; colormap = cgrad([:transparent, :white]; alpha = 0.3))
scatter!(presences; color = :black)
scatter!(bgpoints; color = :red, markersize = 4)Related documentation
PseudoAbsences.PseudoAbsenceGenerator Type
PseudoAbsenceGeneratorAbstract type to which all of the pseudo-absences generator types belong. Note that the pseudo-absence types are singleton types, and the arguments are passed when generating the pseudo-absence mask.
sourcePseudoAbsences.WithinRadius Type
WithinRadiusGenerates pseudo-absences within a set radius (in kilometers) around each occurrence. Internally, this relies on DistanceToEvent.
PseudoAbsences.SurfaceRangeEnvelope Type
SurfaceRangeEnvelopeGenerates pseudo-absences at random within the geographic range covered by actual occurrences. Cells with presences cannot be part of the background sample.
sourcePseudoAbsences.RandomSelection Type
RandomSelectionGenerates pseudo-absences at random within the layer. The full extent is considered, and cells with a true value cannot be part of the background sample.
sourcePseudoAbsences.pseudoabsencemask Function
pseudoabsencemask(::Type{RandomSelection}, presences::SDMLayer{Bool})Generates a mask for pseudo-absences using the random selection method. Candidate cells for the pseudo-absence mask are (i) within the bounding box of the layer (use SurfaceRangeEnvelope to use the presences bounding box), and (ii) valued in the layer.
pseudoabsencemask(::Type{SurfaceRangeEnvelope}, presences::SDMLayer{Bool})Generates a mask from which pseudo-absences can be drawn, by picking cells that are (i) within the bounding box of occurrences, (ii) valued in the layer, and (iii) not already occupied by an occurrence
sourcepseudoabsencemask( ::Type{DistanceToEvent}, presences::SDMLayer{Bool}; f = minimum, )Generates a mask for pseudo-absences using the distance to event method. Candidate cells are weighted according to their distance to a known observation, with far away places being more likely. Depending on the distribution of distances, it may be a very good idea to flatten this layer using log or an exponent. The f function is used to determine which distance is reported (minimum by default, but can be any other relevant function).
pseudoabsencemask(::Type{WithinRadius}, presences::SDMLayer{Bool}; distance::Number = 100.0, )Generates a mask for pseudo-absences where pseudo-absences can be within a distance (in kilometers) of the original observation. Internally, this uses DistanceToEvent.
PseudoAbsences.backgroundpoints Function
backgroundpoints(layer::T, n::Int; kwargs...) where {T <: SimpleSDMLayer}Generates background points based on a layer that gives the weight of each cell in the final sample. The additional keywords arguments are passed to StatsBase.sample, which is used internally. This includes the replace keyword to determine whether sampling should use replacement.