Calibration of SDM outputs
The purpose of this vignette is to show how to calibrate the results of an SDM.
using SpeciesDistributionToolkit
using CairoMakieWe will work on the demo data:
X, y = SDeMo.__demodata()
sdm = SDM(PCATransform, NaiveBayes, X, y)
variables!(sdm, ForwardSelection)PCATransform → NaiveBayes → P(x) ≥ 0.302This model returns the following class scores:
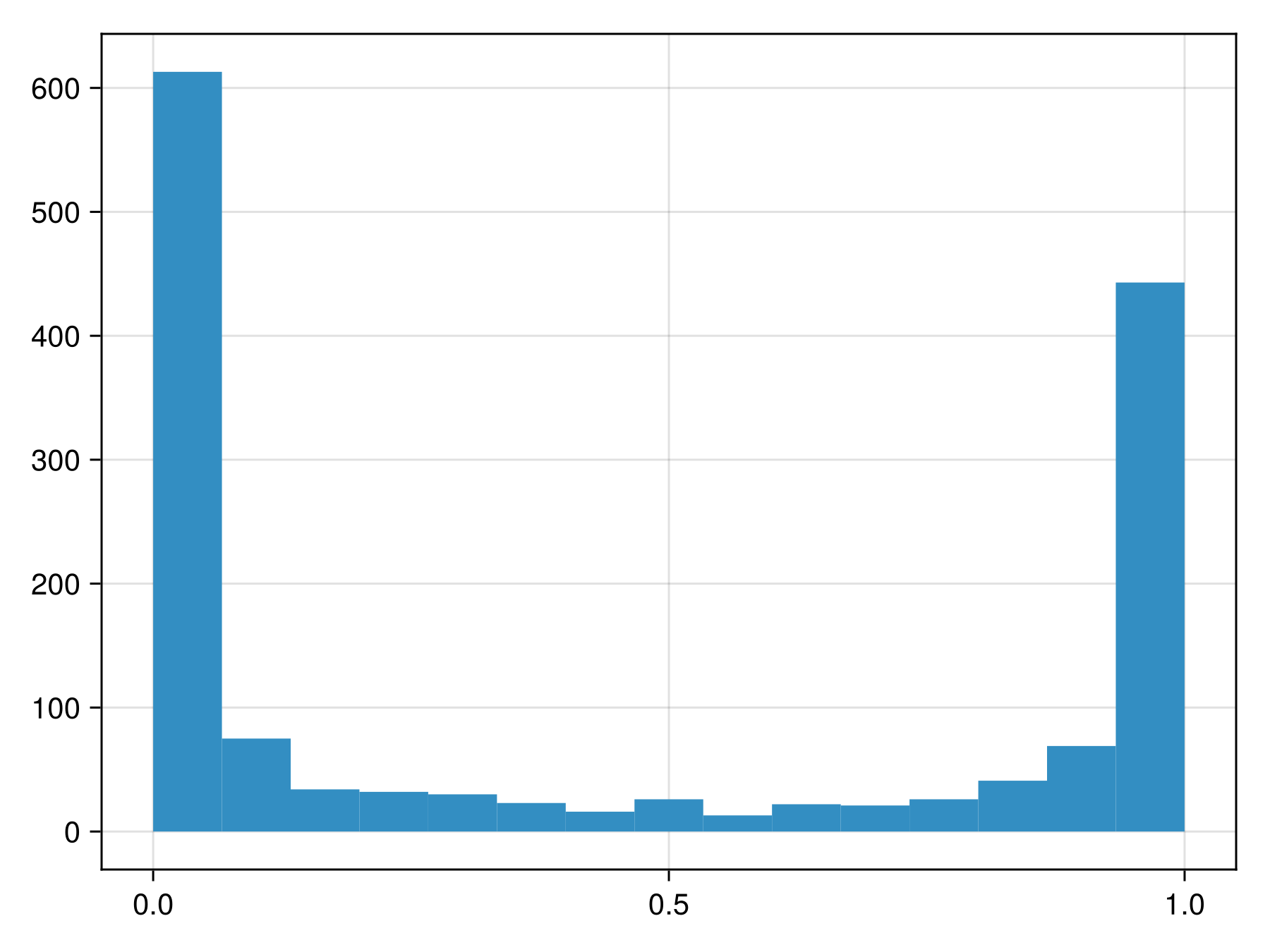
Code for the figure
f = Figure()
ax = Axis(f[1, 1])
hist!(ax, predict(sdm; threshold=false))To figure out whether these are close to actual probabilities, we can look at the reliability curve
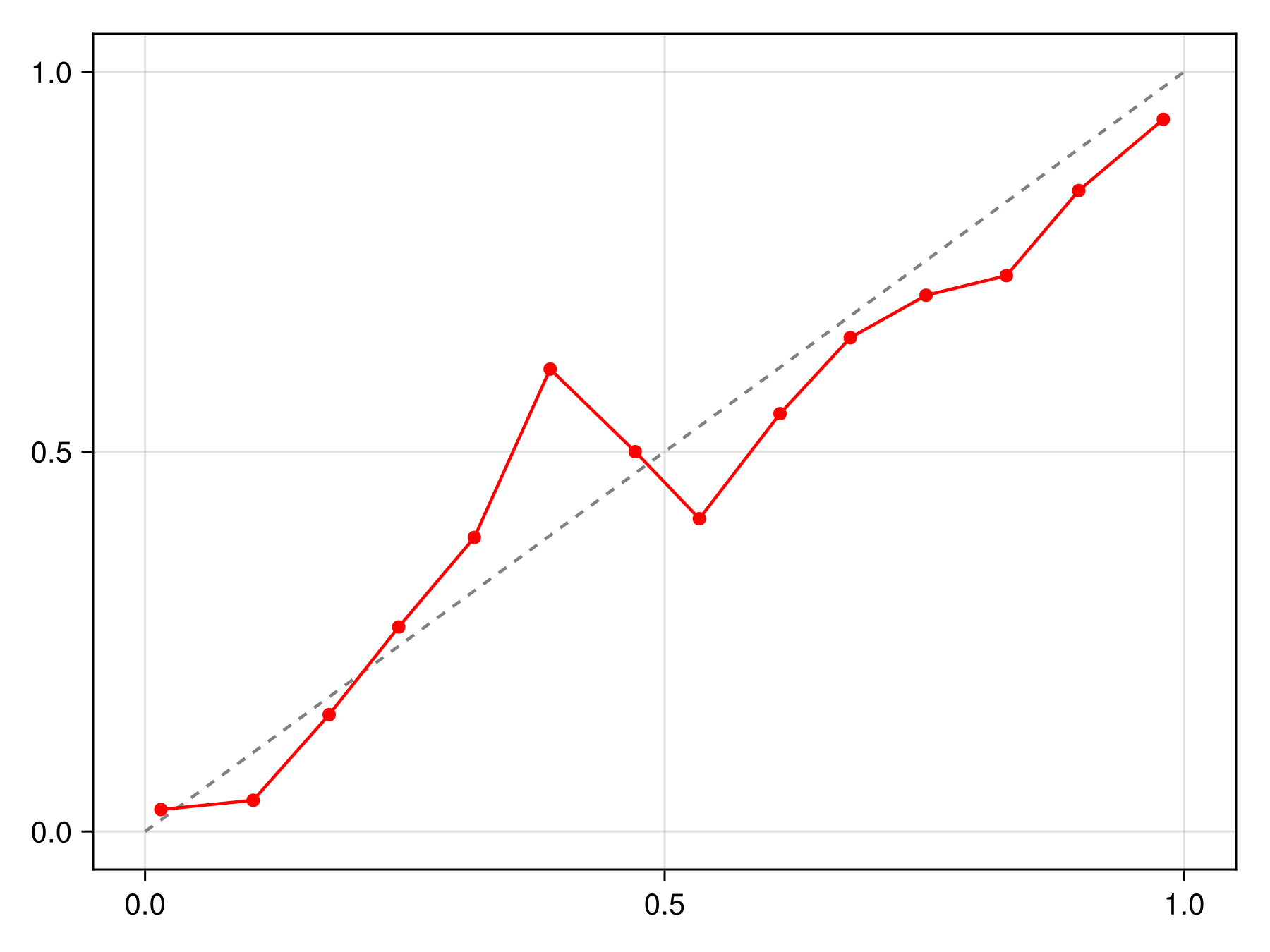
Code for the figure
f = Figure()
ax = Axis(f[1, 1])
lines!(ax, [0, 1], [0, 1], color=:grey, linestyle=:dash)
scatterlines!(ax, reliability(sdm, bins=15)..., color=:red)We can apply different types of calibration functions, such as for example isotonic regression:
C = calibrate(IsotonicCalibration, sdm)IsotonicCalibration(SDeMo.var"#evaluate#PAVA##2"{Vector{Float64}, Vector{Float64}}([0.025179856115107913, 0.048, 0.13333333333333333, 0.16216216216216214, 0.3076923076923077, 0.40625, 0.5000000000000001, 0.5714285714285714, 0.6785714285714286, 0.7380952380952381, 0.7777777777777778, 0.868421052631579, 0.8977272727272727, 0.9414893617021277], [0.010446841702334862, 0.059153266991951135, 0.14602746759462176, 0.19011707586999216, 0.2708963211682988, 0.3110931019922129, 0.38464467093945615, 0.5604360184813539, 0.6927670780514079, 0.7706322760279891, 0.8548092612242976, 0.8965091421785246, 0.9418574850997998, 0.9863507703735425, 0.9863507703735425]))The calibration can be applied by passing it to the correct function, which returns a function to correct a point prediction:
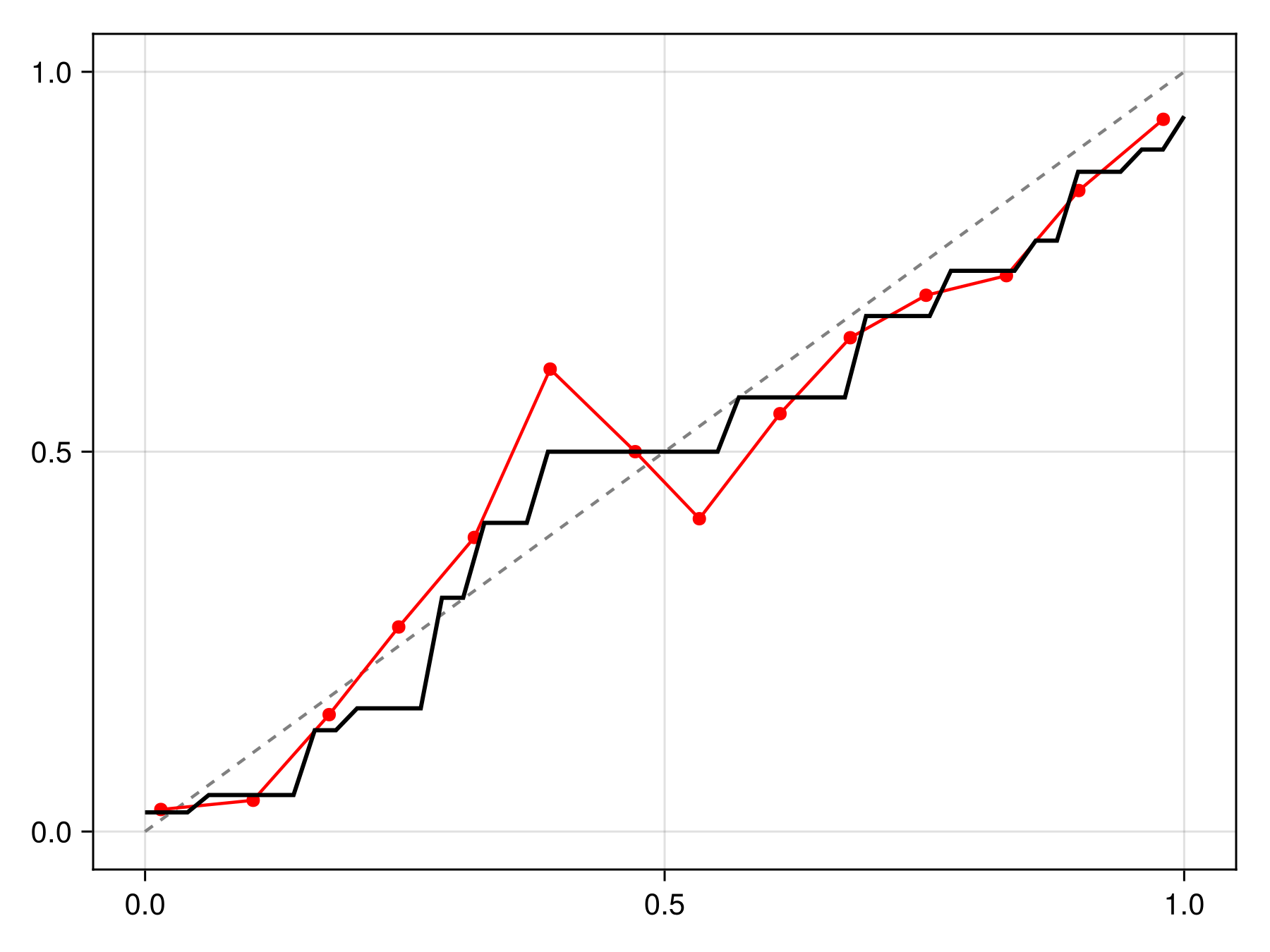
Code for the figure
f = Figure()
x = LinRange(0.0, 1.0, 50)
ax = Axis(f[1, 1])
lines!(ax, [0, 1], [0, 1], color=:grey, linestyle=:dash)
scatterlines!(ax, reliability(sdm, bins=15)..., color=:red)
lines!(ax, x, correct(C).(x), color=:black, linewidth=2)We can check that this calibration is indeed making the model more reliable compared to the initial version:
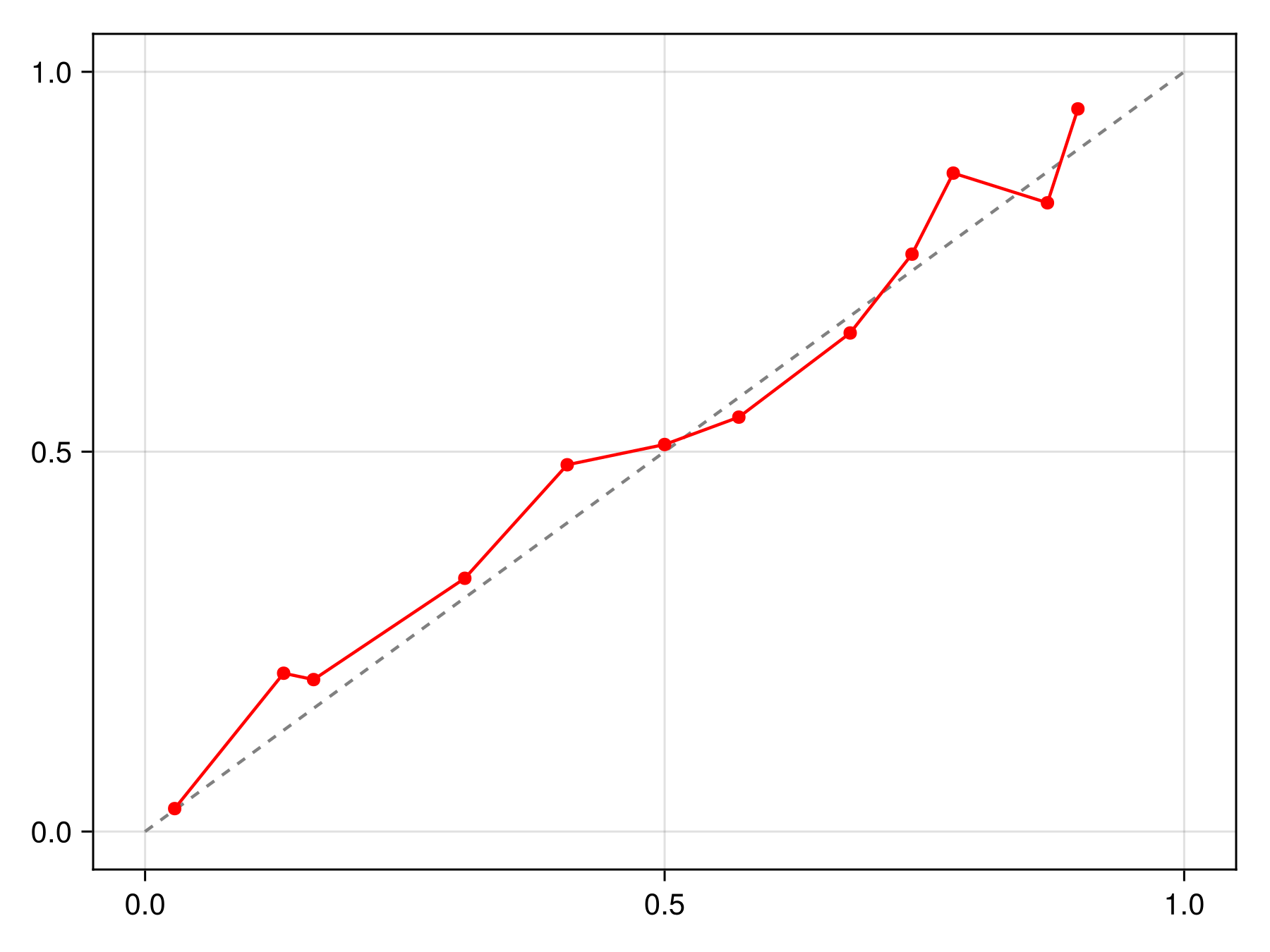
Code for the figure
f = Figure()
x = LinRange(0.0, 1.0, 50)
ax = Axis(f[1, 1])
lines!(ax, [0, 1], [0, 1], color=:grey, linestyle=:dash)
scatterlines!(ax, reliability(sdm, bins=15, link=correct(C))..., color=:red)It is possible to pass a sample keyword to calibrate to only use a series of training points for calibration. This allows to use multiple samples to estimate the calibration function:
samples = first.(bootstrap(sdm))
C = [calibrate(IsotonicCalibration, sdm; samples=s) for s in samples]
cfunc = correct(C)#correct##2 (generic function with 1 method)The correction function will then average the results for each calibration to return the final probability:
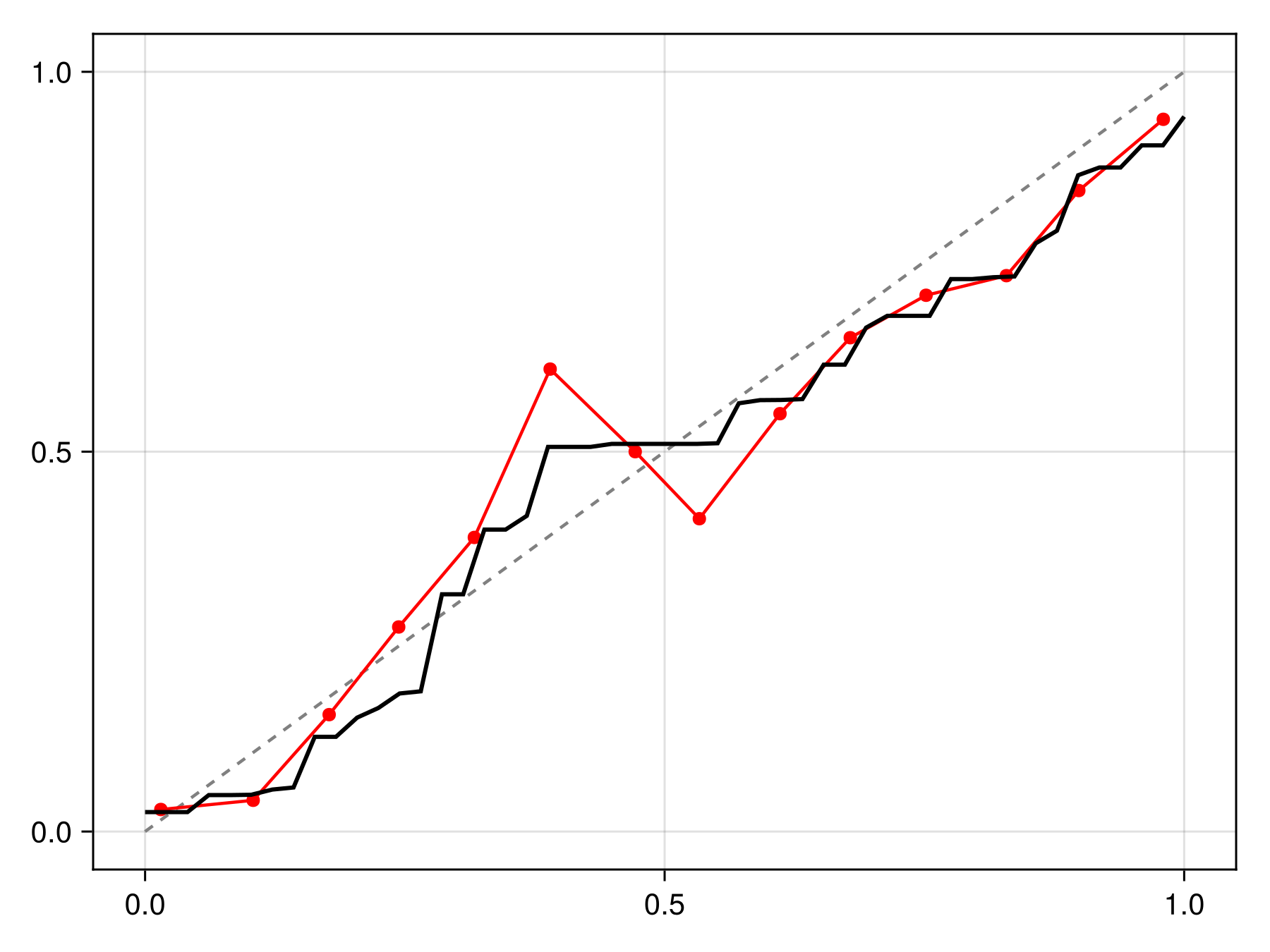
Code for the figure
f = Figure()
x = LinRange(0.0, 1.0, 50)
ax = Axis(f[1, 1])
lines!(ax, [0, 1], [0, 1], color=:grey, linestyle=:dash)
scatterlines!(ax, reliability(sdm, bins=15)..., color=:red)
lines!(ax, x, cfunc.(x), color=:black, linewidth=2)The package also implements Platt's calibration with a fast algorithm, which is appropriate when the relationship between scores and probabilities is sigmoid.