Introduction
Ecosystems are, in large part, constructed by the interactions within them — organisms interact with one-another and with their environment, either directly or indirectly. Interactions between individuals, populations, and species create networks of interactions that drive ecological and evolutionary dynamics and maintain the coexistence, diversity, and functioning of ecosystems (Delmas et al. 2018; Landi et al. 2018; Albrecht et al. 2018). Species interaction networks underpin our understanding of numerous ecological processes (Pascual and Dunne 2006; Heleno et al. 2014). Yet, even basic knowledge of species interactions (like being able to list them, or guess which ones may exist) remains one of the most severe biodiversity shortfalls (Hortal et al. 2015), in large part due to the tedious, time-consuming, and expensive process of collecting species interaction data. Comprehensively sampling every possible interaction is not feasible given the sheer number of species on Earth, and the data we can collect about interactions tend to be biased and noisy (de Aguiar et al. 2019). This is then compounded as species interactions are typically measured as a binary variable (present or absent) even though it is evident interactions are not all-or-nothing. Empirically we know species interactions occur probabilistically due to variation in species abundances in space and time (Poisot, Stouffer, and Gravel 2015). Different types of interactions vary in their intrinsic predictability (e.g. some fungal species engage in opportunistic saprotrophy (Smith et al. 2017), obligate parasites are more deterministic in their interactions than facultative parasites (Poisot et al. 2013; Luong and Mathot 2019)). In addition to this variance in predictability, networks from different systems are structured by different mechanisms.
Still, like all of Earth’s systems, species interaction networks have entered their “long now” (Carpenter 2002), where anthropogenic change will have long-term, low-predictability consequences (Burkle, Marlin, and Knight 2013) for our planet’s ecology. Therefore, our field needs a roadmap towards models that enable prediction (for the present) and forecasting (for the future) of species interactions and the networks they form, and which accounts for their spatial and temporal variation (McCann 2007; Seibold et al. 2018). As an example, in disease ecology, predicting potential hosts of novel disease (recently notably the search for wildlife hosts of betacoronaviruses; Becker et al. 2020; Wardeh, Baylis, and Blagrove 2021) has received much attention. Network approaches have been used for the prediction of risk and dynamics of dengue (Zhao et al. 2020), Chagas disease (Rengifo-Correa et al. 2017), Rickettsiosis (Morand et al. 2020), Leishmaniasis (Stephens 2009), and a myriad infectious diseases in livestock and wildlife (Craft 2015). Additionally, prediction of interaction networks is a growing imperative for next-generation biodiversity monitoring, requiring a conceptual framework and a flexible set of tools to predict interactions that is explicitly spatial and temporal in perspective (Edwards et al. 2021; Magioli and Ferraz 2021; Zhang and He 2021). Developing better models for prediction of these interactions will rely on integration of data from many sources, and the sources for this data may differ depending on the type of interaction we wish to predict (Gibb et al. 2021).
Interactions between species can be conceptualised in a multitude of ways (mutualistic vs. antagonistic, strong vs. weak, symmetric vs. asymmetric, direct vs. indirect) (Jordano 2016a; Morales-Castilla et al. 2015). What is common among definitions of species interactions is that at least one of the species is affected by the presence of another (Morales-Castilla et al. 2015). Networks can be used to represent a variety of interaction types, including: unipartite networks: where each species can be linked to other species (often food webs), bipartite networks: where there are two pools of species and all interactions occur between species in each pool (typically used for pairwise interactions; e.g. hosts and parasites), and k-partite networks,: which expand to more than two discrete sets of interacting species (e.g., some parasitoid webs, seed dispersal networks, and pollination networks (Pocock, Evans, and Memmott 2012)).
Methods for predicting interactions between species exist, but at the moment are difficult to generalise as they are typically based around a single mechanism at a single scale: position in the trophic niche (Gravel et al. 2013; Petchey et al. 2008), phylogenetic distance (Pomeranz et al. 2018; Elmasri et al. 2020), functional trait matching (Bartomeus et al. 2016), interaction frequency (Weinstein and Graham 2017; Vázquez, Morris, and Jordano 2005), or other network properties (Terry and Lewis 2020; Stock et al. 2017). Species interaction networks, as we observe them on Earth today, are the product of ecological and evolutionary mechanisms interacting across spatial, temporal and organisational scales. The interwoven nature of these processes imposes structure on biodiversity data which is invisible when examined only through the lens of a single scale, however machine learning (ML) methods have enormous potential to find this structure in data (Desjardins-Proulx, Poisot, and Gravel 2019), and have the potential to be used together with mechanistic models in order to make prediction of ecological dynamics more robust (Rackauckas et al. 2020).
Here we use a case study to show how machine-learning models (specifically a deep neural network) can enable prediction of species interactions: we construct a metaweb of host-parasite interactions across space, using predictors extracted from empirical data and accounting for the structure of co-occurrence between species. We use this case study to illustrate a roadmap for improving predictions using open data and ML methods; specifically, we focus on how emerging tools from ML can be used to deliver more accurate and more efficient predictions of ecological systems, and how the potential of these approaches will be magnified with increased data access. We then provide a non-exhaustive primer on the literature on interaction prediction, and identify the tools and methods most suited for the future of interaction network prediction models, covering the spatial, temporal, and climatic dimensions of network prediction (Burkle and Alarcon 2011). Both the case study and primer are largely geared towards binary (interactions are either present or absent) networks; there are limitations in data and tools that make it a more reasonable starting approach. First, most ecological networks do not have estimates of interaction strength, and particularly not estimates that are independent from relative abundances. Second, the methodological toolkit to analyse the structure of networks is far more developed for binary interactions (Delmas et al. 2018), meaning that the predictions of binary interactions can be more readily interpreted.
We argue that adopting a more predictive approach to complex ecological systems (like networks) will establish a positive feedback loop with our understanding of these systems (Houlahan et al. 2017): the tasks of understanding and predicting are neither separate nor opposed (Maris et al. 2017); instead, ML tools have the ability to capture a lot of our understanding into working assumptions, and comparing predictions to empirical data gives us better insights about how much we ignore about the systems we model (see for example Borowiec et al. 2021, who provide an overview of deep learning techniques and concepts in ecology and evolution). Although data on species interaction networks are currently limited in the size and spatial coverage, machine learning approaches have a demonstrated track record of revealing the “unreasonable effectiveness” of data (Halevy, Norvig, and Pereira 2009); we argue that with a clear roadmap guiding the use of these methods, the task of predicting species interaction networks will become more attainable.
A case study: deep learning of spatially sparse host-parasite interactions
The premise of this manuscript is that we can predict interactions between species. In this section we provide a proof-of-concept, where we use data from Hadfield et al. (2014) describing 51 host-parasite networks sampled across space. In this data, as in most spatially distributed ecological networks, not all species co-occur across sites. As a direct consequence there are pairs of species that may or may not be able to interact for which we have no data; furthermore there are pairs of species that may interact, but have only been documented in a single location where the interaction was not detected. In short, there are ecological reasons to believe that a number of negative associations in the metaweb (sensu J. Dunne 2006) are false negatives.
Without any species-level information, we resort to using both co-occurrence and known interactions to predict novel interactions. To do this we (i) extract features (equivalent to explanatory variables in a statistical model) for each species based on co-occurrence, (ii) use these features to train an artificial neural network to predict interactions, and (iii) apply this classifier (an algorithm that assigns a categorical output based on input features) to the original features to predict potential interactions across the entire species pool. Machine learning relies on a lexicon that shares some terms with statistics, albeit with different meaning; we expand on the precise meanings in the “How to validate a predictive model” section below. The outputs of the analysis are presented in fig. 1, and the code to reproduce it is available at https://osf.io/6jp4b/; the entire example was carried out in Julia 1.6.2 (Bezanson et al. 2017), using the Flux machine learning framework (Innes 2018).
We first aggregate all species into a co-occurrence matrix A which represents whether a given pair of species (i, j) was observed coexisting across any location. We then transform this co-occurrence matrix A via probabilistic PCA (Tipping and Bishop 1999) and use the first 15 values from this PCA space as the features vector for each species i. For each pair of (host, parasite) species (i, j), we then feed the features vectors (vi, vj) into a neural network. The neural network uses four feed-forward layers (each layer is independent from the one before and after); the first layer uses the RELU activation function (which ignores input below a threshold), the rest use a σ function (which transforms linear activation energies into logistic responses). All layers have appropriate dropout rates (in order to avoid over-fitting, only a fraction of the network is updated on each iteration: 1 − 0.8 for the first layer, 1 − 0.6 for the subsequent ones). This produces an output layer with a single node, which is the probability-score for interaction between species i and j.
We then train (equivalent to fit) this neural network by dividing the original dataset into testing and training sets (split 80-20 for training and testing respectively). During the training of this neural network (using the ADAM optimiser), the 5 × 104 batches of 64 items used for training were constrained to have at least 25% of positive interactions, as Poisot, Ouellet, et al. (2021) show slightly inflating the dataset with positive interactions enables us to counterbalance sampling biases. Furthermore, setting a minimum threshold of response balance is an established approach for datasets with strong biases (Lemaître, Nogueira, and Aridas 2017). Validating this model on the test data shows our model provides highly effective prediction of interactions between pairs of species not present in the training data (fig. 1). The behaviour of the model was, in addition, checked by measuring the training and testing loss (difference between the actual value and the prediction, here using mean-squared error) and stopping well before they diverged (to avoid overfitting).
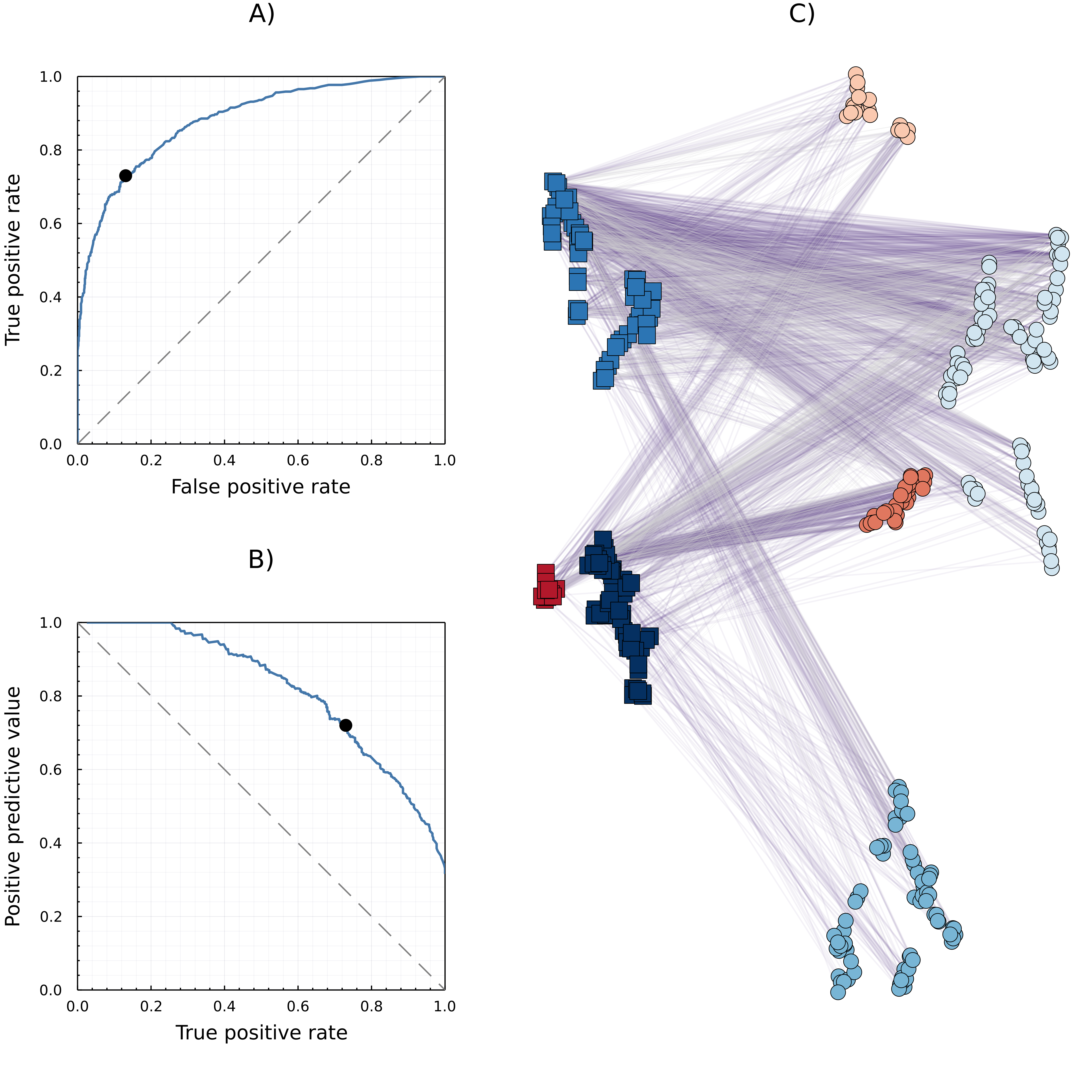
This case study shows that a simple neural network can be very effective in predicting species interactions even without additional species-level data. Applying this model to the entire dataset (including species pairs never observed to co-occur) identified 1546 new possible interactions – 746 (48%) of which were between pairs of species for which no co-occurrence was observed in the original dataset. This model reaches similar levels of predictive efficacy as previous studies that use far more species-level data and mechanistic assumptions (Gravel et al. 2013), which serves to highlight the potential for including external sources of data for improving our prediction of interaction networks even further. For example, Krasnov et al. (2016) collected traits data for this system that could be added to the model, in addition or in substitution to latent variables derived from observed interactions.
Predicting species interaction networks across space: challenges and opportunities
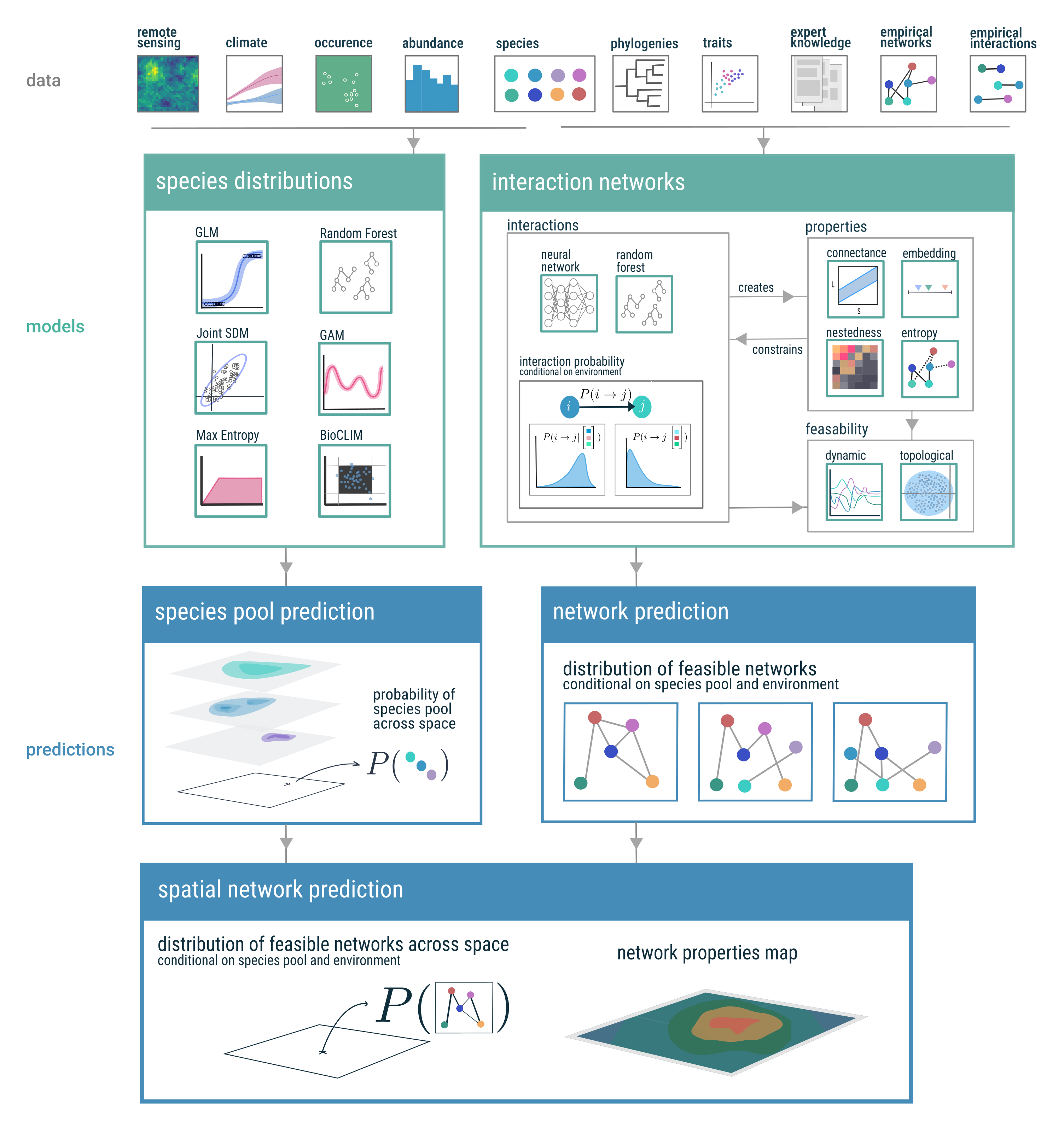
Here we present a conceptual roadmap (fig. 2) which shows a conceptual path from data to prediction of species interaction networks, incorporating several modelling frameworks. We envisage this roadmap to be one conceptual path toward incorporating space in to our prediction of interaction networks, and developing spatially explicit models of networks and their properties. In the following sections we discuss the challenges and opportunities for this path forward, and highlight two specific areas where it can have a strong impact: the temporal forecasting of species interaction networks structure, and the use of predicted networks for applied ecology and conservation biology.
Challenges: constraints on predictions
Ecological network data are scarce and hard to obtain
At the moment, prediction of species interactions is made difficult by the limited availability of data. Although we have seen a growth in species occurrence data, this growth is much slower for ecological interactions because species interactions are challenging to sample comprehensively (Bennett, Evans, and Powell 2019; Jordano 2016b) and sampling methodology has strong effects on the resulting data (de Aguiar et al. 2019). In turn, the difficulty of sampling interactions can lead to biases in our understanding of network structure (de Aguiar et al. 2019). This knowledge gap has motivated a variety of approaches to deal with interactions in ecological research based on assumptions that do not always hold, such as the assumption that co-occurrence is equivalent to meaningful interaction strength (Blanchet, Cazelles, and Gravel 2020). Spatial biases in data coverage are prevalent at the global scale (with South America, Africa and Asia being under-represented) and different interaction types show biases towards different biomes (Poisot, Bergeron, et al. 2021). These “spatial gaps” serve as a limitation to our ability to confidently make predictions when accounting for real-world environmental conditions, especially in environments for which there are no analogous data.
Further, empirical estimation of interaction strength is highly prone to bias as existing data are usually summarised at the taxonomic scale of the species or higher, thereby losing information that differentiates the strength in per-individual interactions from the strength of a whole species interaction (Wells and O’Hara 2013). Empirical estimations of interaction strength are still crucial (Novak and Wootton 2008), but are a hard task to quantify in natural communities (Wootton 1997; Sala and Graham 2002; Wootton and Emmerson 2005), especially as the number of species composing communities increases, compounded by the possibility of higher-order interactions or non-linear responses in interactions (Wootton and Emmerson 2005). Further, interaction strength is often variable and context dependent and can be influenced by density-dependence and spatio-temporal variation in community composition (Wootton and Emmerson 2005).
Powerful predictive tools work better on large data volumes
This scarcity of data limits the range of computational tools that can be used by network ecologists. Most deep learning methods, for instance, are very data expensive. The paucity of data is compounded by a collection of biases in existing datasets. Species interaction data are typically dominated by food webs, pollination, and host-parasite networks (Ings et al. 2009; Poisot et al. 2020). This could prove to be a limiting factor when trying to understand or predict networks of underrepresented interaction types or when trying to integrate networks of different types (Fontaine et al. 2011), especially given their inherent structural variation (Michalska-Smith and Allesina 2019). This stresses the need for an integrated, flexible, and data-efficient set of computational tools which will allow us to predict ecological networks accurately from existing and imperfect datasets, but also enable us to perform model validation and comparison with more flexibility than existing tools. We argue that fig. 1 is an example of the promise of these tools even when facing datasets of small size. The ability to extract and engineer features also serves to bolster our predictive power. Although it may be tempting to rely on approaches like bootstrapping to estimate the consistency of the predictions, we are confronted with the issues of low data volume and data bias—that we are more likely to observe interactions between some pairs of species (i.e. those that co-occur often, e.g. Cazelles et al. (2015), and those with higher relative abundance, e.g. Vazquez et al. (2009)). This introduces risk in training models on pseudo-replicated data. In short, the current lack of massive datasets must not be an obstacle to prediction; it is an ideal testing ground to understand how little data is sufficient to obtain actionable predictions, and how much we can rely on data inflation procedures to reach this minimal amount.
Scaling-up predictions requires scaled-up data
We are also currently limited by the level of biological organisation at which we can describe ecological networks. For instance, our understanding of individual-based networks (e.g., M. S. Araújo et al. 2008; Tinker et al. 2012) is still in its infancy (Guimarães 2020) and acts as a resolution-limit. Similarly, the resolution of environmental (or landscape) data also limits our ability to predict networks at small scales, although current trends in remote sensing would suggest that this will become less of a hindrance with time (Makiola et al. 2020). Ecosystems are a quintessential complex-adaptive-system (Levin 1998) with a myriad of processes at different spatial, temporal, and organisational scales that influence and respond to one another. Understanding how the product of these different processes drive the properties of ecosystems across different scales remains a central challenge of ecological research, and we should strive to work on methods that will integrate different empirical “snapshots” of this larger system.
Opportunities: an emerging ecosystem of open tools and data
Data are becoming more interoperable
The acquisition of biodiversity and environmental data has tremendously increased over the past decades thanks to the rise of citizen science (J. L. Dickinson, Zuckerberg, and Bonter 2010) and of novel technology (Stephenson 2020), including wireless sensors (Porter et al. 2005), next-generation DNA sequencing (Creer et al. 2016), and remote sensing (Skidmore and Pettorelli 2015; Lausch et al. 2016). Open access databases, such as GBIF (for biodiversity data), NCBI (for taxonomic and genomics data), TreeBASE (for phylogenetics data), CESTE (Jeliazkov et al. 2020) (for metacommunity ecology and species traits data), and WorldClim (for bioclimatic data) contain millions of data points that can be integrated to monitor and model biodiversity at the global scale. For species interactions data, at the moment Mangal is the most comprehensive open database of published ecological networks (Poisot et al. 2016), and GloBI is an extensive database of realised and potential species interactions (Poelen, Simons, and Mungall 2014). Developing standard practices in data integration and quality control (Kissling et al. 2018) and in next-generation biomonitoring (NGB; Makiola et al. 2020) would improve our ability to make reliable predictions of ecosystem properties on increasing spatial and temporal scales. The advancement of prediction techniques coupled with a movement towards standardising data collection protocols (e.g. Pérez-Harguindeguy et al. (2013) for plant functional traits) and metadata (e.g. DarwinCore)—which facilitates interoperability and integration of datasets—as well as a growing interest at the government level (Scholes et al. 2012) paints a positive picture for data access and usability in the coming years.
Machine learning tools are becoming more accessible
This effort is also supported by a thriving ecosystem of data sources and novel tools. ML methods can often be more flexible and perform better than classical statistical methods, and can achieve a very high level of accuracy in many predictive and classification tasks in a relatively short amount of time (e.g., Cutler et al. 2007; Krizhevsky, Sutskever, and Hinton 2017). Increasing computing power combined with recent advances in machine learning techniques and applications shows promise in ecology and environmental science (see Christin, Hervet, and Lecomte (2019) for an overview). Moreover, ongoing developments in deep learning are aimed at improvement in low-data regimes and with unbalanced datasets (Antoniou, Storkey, and Edwards 2018; Chawla 2010). Considering the current biases in network ecology (Poisot, Bergeron, et al. 2021) and the scarcity of data of species interactions, the prediction of ecological networks will undoubtedly benefit from these improvements. Machine learning methods are emerging as the new standard in computational ecology in general (Olden, Lawler, and Poff 2008; Christin, Hervet, and Lecomte 2019), and in network ecology in particular (Bohan et al. 2017), as long as sufficient, relevant data are available. Many studies have used machine learning models specifically with ecological interactions. Relevant examples include species traits used to predict interactions and infer trait-matching rules (Desjardins-Proulx et al. 2017; Pichler et al. 2020), automated discovery of food webs (Bohan et al. 2011), reconstruction of ecological networks using next-generation sequencing data (Bohan et al. 2017), and network inference from presence-absence data (Sander, Wootton, and Allesina 2017). As many ecological and evolutionary processes underlie species interactions and the structure of their ecological networks (e.g., Vazquez et al. 2009; Segar et al. 2020), it can be difficult to choose relevant variables and model species interactions networks explicitly. A promising application of machine learning in natural sciences is Scientific-Machine Learning (SciML), a framework that combines machine learning with mechanistic models (Chuang and Keiser 2018; Rackauckas et al. 2020).
A primer on predicting ecological networks
Within the constraints outlined in the previous section, we now provide a primer on the background concepts necessary to build predictive models of species interaction networks, with a focus on using machine learning approaches in the modelling process. As fig. 2 illustrates, this involves a variety of numerical and computational approaches; therefore, rather than an exhaustive summary, we aim to convey a high-level understanding that translates the core concepts into their application to ecological networks.
Models
What is a predictive model?
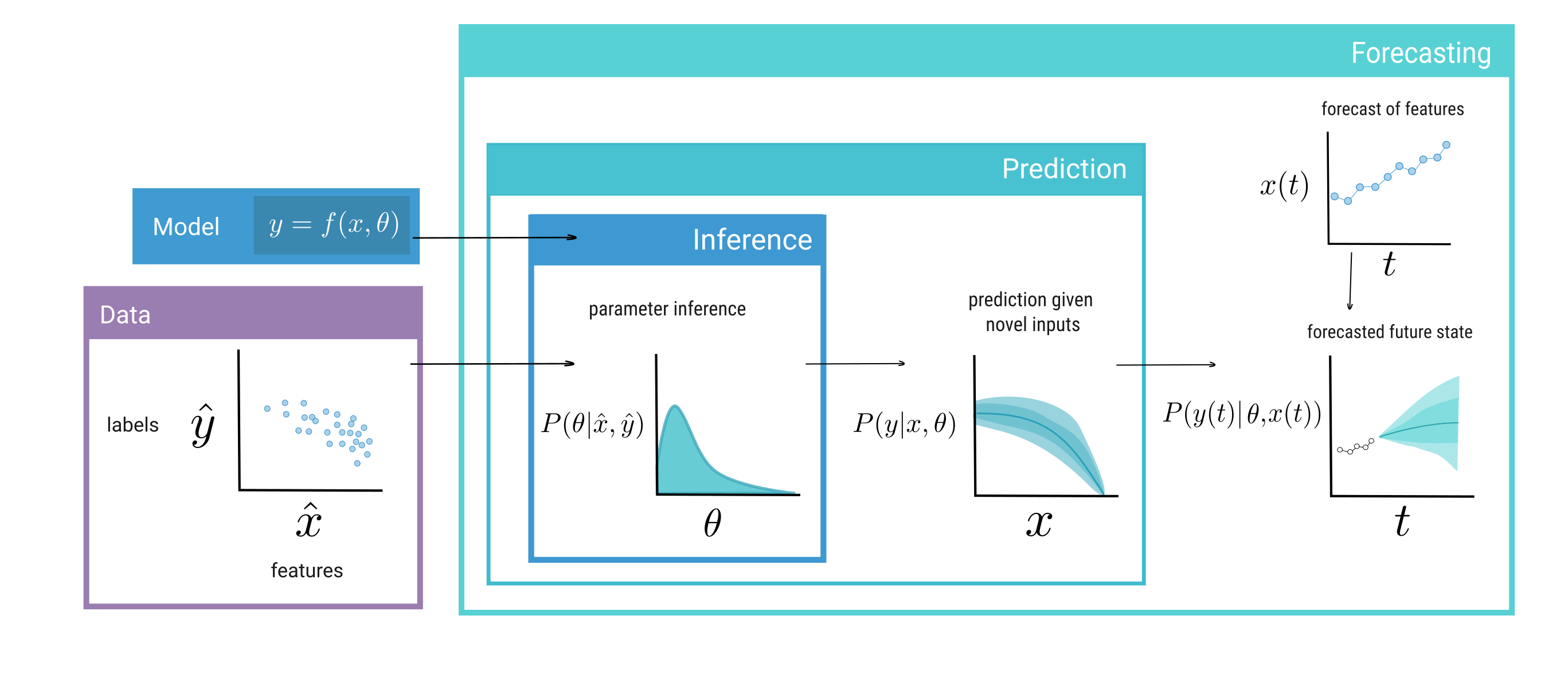
Models are used for many purposes, and the term “model” itself embodies a wide variety of meanings in scientific discourse. All models can be thought of as a function, f, that takes a set of inputs x (also called features, descriptors, or independent variables) and parameters θ, and maps them to predicted output states y (also called label, response, or dependent variable) based on the input to the model: y = f(x, θ).
A given model f can be used for either descriptive or predictive purposes. Many forms of scientific inquiry are based around using models descriptively, a practice also called inference, the inverse problem, fitting a model, or training a model (Stouffer 2019). In this context, the goal of using a model is to estimate the parameters, θ, that best explain a set of empirical observations, {x̂, ŷ}. In some cases, these parameter values are themselves of interest (e.g., the strength of selection, intrinsic growth rate, dispersal distance), but in others cases, the goal is to compare a set of competing models f1, f2, … to determine which provides the most parsimonious explanation for a dataset. The quantitative representation of “effects” in these models—the influence of each input on the output—is often assumed to be linear, and within the frequentist world-view, the goal is often to determine if the coefficient corresponding with an input is non-zero to determine its “significance” (often different from its ecological relevance; Martínez-Abraín 2008) in influencing the outcome.
Models designed for inference have utility—descriptive models of networks can reveal underlying mechanisms that structure ecological communities, given a proper null model (Connor, Barberán, and Clauset 2017). However, in order for ecology to develop as a predictive science (Evans, Norris, and Benton 2012), interest has grown in developing models that are used not just for description of data, but also for prediction. Predictive models are based in the forward problem, where the aim is to predict new values of the output y given an input x and our estimate value of θ (Stouffer 2019). Because the forward problem relies on an estimate of θ, then, the problem of inference is nested within the forward problem (fig. 3): working towards a predictive view of ecological networks will give us the needed tools to further our understanding of them.
What do you need to build a predictive model?
To build a predictive model, one needs the following: first, data, split into features x̂ and labels ŷ (fig. 3). Second, a model f, which maps features x to labels y as a function of parameters θ, i.e. y = f(x, θ). Third, a loss function L(ŷ, y), which describes how far a model’s prediction y is from an empirical value ŷ. Lastly, priors on parameters, P(θ), which describe the modeller’s a priori belief about the value of the parameters; rather than making an analysis implicit, specifying priors has the merit of making the modeller’s assumptions explicit, which is a most desirable feature when communicating predictions to stakeholders (Spiegelhalter et al. 2000). Often an important step before fitting a model is feature engineering: adjusting and reworking the features to better uncover feature-label relationships (Kuhn and Johnson 2019). This can include projecting the features into a lower dimensional space, as we did through a probabilistic PCA in the case study, or removing the covariance structure using a Whitening approach. Then, when a model is fitted (synonymous with parameter inference or the inverse problem, see fig. 3), a fitting algorithm attempts to estimate the values of θ that minimises the mean value of loss function L(ŷ, y) for all labels ŷ in the provided data Y. In a Bayesian approach, this typically relys on drawing candidate parameter values from priors and applying some form of sampling to generate a posterior estimate of parameters, P(θ|x̂, ŷ). In the training of neural networks, this usually involves some form of error back-propagation across the edges in order to tune their weights, and the biases of each nodes.
How do we validate a predictive model?
After we fit a model, we inevitably want to see how “good” (meaning, “fit for purpose”) it is. This process can be divided into two parts: (i)) model selection, where the modeller chooses from a set of possible models and (ii) model assessment, where the modeller determines the performance characteristics of the chosen model (Hastie, Tibshirani, and Friedman 2009).
In the context of model selection, a naïve initial approach is to simply compute the average error between the model’s prediction and the true data we have, and choose the model with the smallest error—however this approach inevitably results in overfitting. One approach to avoid overfitting is using information criteria (e.g., AIC, BIC, MDL) based around the heuristic that good models maximise the ratio of information provided by the model to the number of parameters it has. However, when the intended use-case of a model is prediction the relevant form of validation is predictive accuracy, which should be tested with cross-validation. Cross-validation methods divide the original dataset into two—one which is used to fit the model (called the training set) and one used to validate its predictive accuracy on the data that it hasn’t “seen” yet (called the test set) (Bishop 2006). This procedure is often repeated across different test and training subdivisions of the dataset (either picked randomly or stratified by some criteria, like balance between positive and negative interactions in the case study) to determine the uncertainty associated with our measurement due to our choice of test and training sets (Arlot and Celisse 2010), in the same conceptual vein as data bootstrapping: the mean value of the validation metric gives an overall estimate of its performance, and the variance around this mean represents the effect of using different data for training and testing. In a robust model/dataset combination, we expect this variance to be low, although there are no prescriptive guidelines as to how little variance is acceptable; the choice of whether to use a model is often left to the best judgement of the modeller.
We still have to define what predictive accuracy means in the context of interaction network prediction. In the proof-of-concept, we used a neural-network to perform binary classification by predicting the presence/absence of an interaction between any two species. There are two ways for the model to be right: the model predicts an interaction and there is one (a true positive (TP)), or the model predicts no interaction and there isn’t one (a true negative (TN)). Similarly, there are two ways for the model to be wrong: the model predicts an interaction which does not exist (a false positive (FP)), or the model predicts no interaction but it does exist (a false negative (FN)).
A naïve initial approach to measure how well a model does is accuracy, i.e. the proportion of values it got correct. However, consider what we know about interaction networks: they are often very sparse, with connectance usually below a third (Cohen, Briand, and Newman 1990). If we build a model that always guesses there will be no interaction between two species, it will be correct in the majority of cases because the majority of potential interactions in a network typically do not exist. Therefore this “empty-matrix” model would always have an accuracy of 1 − C, where C is the observed connectance, which would almost always be greater than 50%. Understanding model performance within sensitivity-specificity space may be more informative, where sensitivity evaluates how good the model is at predicting true interactions (True Positive Rate) and specificity refers to the prediction of true “non-interactions” (True Negative Rate). It must be noted that in ecological networks, there is no guarantee that the “non-interactions” (assumed true negatives) in the original dataset are indeed true negatives (Jordano 2016a, 2016b). This can result in the positive/negative values, and the false omission/discovery being artificially worse, and specifically decrease our confidence in predicted interactions.
In response to the general problem of biases in classifiers, many metrics have been proposed to measure binary-classifiers (Gu, Zhu, and Cai 2009; Drummond and Holte 2006) and are indicative of how well the model performs with regards to some aspect of accuracy, sensitivity, specificity and/or precision (tbl. 1). Ultimately the choice of metric will depend on the intended use of the model: there is not a single definition of “success,” but rather different interpretation of what sources of error are acceptable for a given application.
| Name | Value | Success | Description |
|---|---|---|---|
| Random accuracy | 0.56 | Fraction of correct predictions if the classifier is random | |
| Accuracy | 0.81 | → 1 | Observed fraction of correct predictions |
| Balanced accuracy | 0.80 | → 1 | Average fraction of correct positive and negative predictions |
| True Positive Rate | 0.77 | → 1 | Fraction of interactions predicted |
| True Negative Rate | 0.83 | → 1 | Fraction of non-interactions predicted |
| False Positive Rate | 0.16 | → 0 | Fraction of non-interactions predicted as interactions |
| False Negative Rate | 0.22 | → 0 | Fraction of interactions predicted as non-interactions |
| ROC-AUC | 0.86 | → 1 | Proximity to a perfect prediction (ROC-AUC=1) |
| Youden’s J | 0.60 | → 1 | Informedness of predictions (trust in individual prediction) |
| Cohen’s κ | 0.58 | ≥ 0.5 | |
| Positive Predictive Value | 0.66 | → 1 | Confidence in predicted interactions |
| Negative Predictive Value | 0.89 | → 1 | Confidence in predicted non-interactions |
| False Omission Rate | 0.10 | → 0 | Expected proportion of missed interactions |
| False Discovery Rate | 0.33 | → 0 | Expected proportion of wrongly imputed interactions |
In the machine learning literature, a common way of visualising this extensive list of possible metrics is through the use of ROC (receiver-operating-characteristic; False Positive Rate on the x-axis, and True Positive Rate on the y-axis) and PR (precision-recall; True-Positive-Rate on the x-axis, Positive-predictive-value on the y-axis) curves (see fig. 1). These curves are generated by considering a continuum of thresholds of classifier acceptance, and computing the values of ROC/PR metrics for each value of the threshold. The area-under-the-curve (AUC) is then used as a validation metric and are typically called AUC-ROC (Area-Under-the-Curve Receiver-Operator-Curve) and AUC-PR (Area-Under-the-Curve Precision-Recall) (e.g. ROC-AUC in tbl. 1). These measures have the unstated assumption that the training and testing set are “correct,” or at least correct enough that the number of true/false positive/negatives are meaningful; although should this assumption be true, there would be no need for any predictive approach – but it is a well established fact that machine learning systems are resilient to even relatively high uncertainties in the data (Halevy, Norvig, and Pereira 2009).
Networks and interactions as predictable objects
Why predict networks and interactions at the same time?
Ecological networks are quite sparse, and larger networks tend to get sparser (MacDonald, Banville, and Poisot 2020); in other words, although networks are composed of a set of interactions between species pairs, they also form a much larger set of species pairs that do not interact. If we aim to predict the structure of networks from the “bottom-up”— by considering each pairwise combination of S different species—we are left with S2 interaction values to estimate, a majority of which will be 0. Instead, we can use our existing understanding of the mechanisms that structure ecological networks to whittle down the set of feasible adjacency matrices, thereby reducing the amount of information we must predict, and making the problem of predicting interactions less daunting. The processes that structure ecological networks do not only occur at the scale of interactions—there are also processes at the network level which limit what interactions (or how many) are realistic. The realised structure of a network is the synthesis of the interactions forming the basis for network structure, and the network structure refining the possible interactions—“Part makes whole, and whole makes part” (Levins and Lewontin 1987).
Another argument for the joint prediction of networks and interactions is to reduce circularity and biases in the predictions. As an example, models like linear filtering (Stock et al. 2017) generate probabilities of non-observed interactions existing, but do so based on measured network properties. Some recent models make interaction-level predictions (e.g. Gravel et al. 2019); these are not unlike stacked species distribution models, which are individually fit, but collectively outperformed by joint models or rule-based models (Zurell et al. 2020). By relying on adequate testing of model performance of biases (i.e. optimising not only accuracy, but paying attention to measures like false discovery and false omission rates), and developing models around a feedback loop between network and interaction prediction, it is likely that the quality of the predicted networks will be greatly improved compared to current models.
What network properties should we use to inform our predictions of interactions?
There are many dimensions of network structure (Delmas et al. 2018), yet there are two arguments to support basing network prediction around a single property: connectance (the ratio of actual edges to possible edges in the network). First, connectance is ecologically informative—it relates to resilience to invasion (Baiser, Russell, and Lockwood 2010; Smith-Ramesh, Moore, and Schmitz 2016), can increase robustness to extinction in food webs (J. Dunne, Williams, and Martinez 2002), while decreasing it in mutualistic networks (Vieira and Almeida-Neto 2015), and connectance relates to network stability (Landi et al. 2018). Second, most (if not all) network properties covary with connectance (Poisot and Gravel 2014; J. A. Dunne, Williams, and Martinez 2002).
Within the network science literature, there are numerous methods for predicting edges based on network properties (e.g., block models (Yen and Larremore 2020) based on modularity, hierarchical models (Kawakatsu et al. 2021) based on embedding, etc.). However, in the context of species interaction networks, these properties often covary with connectance. As a result we suggest that using connectance as the primary property of interest is most likely to be practical to formulate at the moment. We have models to estimate species richness over space (Jenkins, Pimm, and Joppa 2013), and because we can predict connectance from species richness alone (MacDonald, Banville, and Poisot 2020), we can then derive distributions of network properties from richness estimates, that can serve to penalise further models that formulate their predictions at the scale of each possible interaction.
How do we predict how species that we have never observed together will interact?
A neutral approach to ecological interactions would assume the probability of an interaction to mirror the relative abundance of both species, and would be unaffected by trait variation (Poisot, Stouffer, and Gravel 2015; Pichler et al. 2020); more accurately, a neutral assumption states that the relative abundances are sufficient to predict the structure of networks, and this view is rather well supported in empirical and theoretical systems (Canard et al. 2012, 2014). However, functional-trait based proxies could enable better predictions of ecological interactions (Cirtwill and Eklöf 2018; Cirtwill et al. 2019; Bartomeus et al. 2016; Bartomeus 2013). Selection on functional traits could cause interactions to be conserved at some evolutionary scales, and therefore predictions of interaction could be informed by phylogenetic analyses (Davies 2021; Elmasri et al. 2020; Gómez, Verdú, and Perfectti 2010). Phylogenetic matching in bipartite networks is consistent across scales (Poisot and Stouffer 2018), even in the absence of strong selective pressure (Coelho, Rodrigues, and Rangel 2017).
A separate family of methods are based on network embedding (as in the proof-of-concept). A network embedding projects each node of the network into a lower-dimensional latent space. Previous explorations of the dimensionality of food webs have revealed that a reduced number of dimensions (7) was sufficient to capture most of their structure (Eklöf et al. 2013); however, recent quantifications of the complexity of the embedding space of bipartite ecological networks found a consistent high complexity (Strydom, Dalla Riva, and Poisot 2021), suggesting that the precise depth of embedding required may vary considerably across systems. Embeddings enables us to represent the structure of a network, which previously required the S2 dimensions of an adjacency matrix, with a smaller number of dimensions. The position of each node in this lower dimensional space is then treated as a latent measurement corresponding to the role of that species in the network (e.g. Poisot, Ouellet, et al. 2021, where a network of about 1500 species was most accurately described using 12 dimensions). Species close together in the latent space should interact with similar set of species (Rossberg et al. 2006; Rohr et al. 2010). However, these models are sensitive to sampling biases as they are limited to species for which there is already interaction data, and as a result a methodological breakthrough is needed to extend these models to species for which there is little or no interaction data.
How do we quantify interaction strength?
Species interaction networks can also be used as a means to quantify and understand interaction strength. Interaction strength, unlike the qualitative presence or absence of an interaction, is a continuous measurement which attempts to quantify the effect of one species on another. This results in weighted networks representing different patterns of ‘flows’ between nodes – which can be modelled in a variety of ways (Borrett and Scharler 2019). Interaction strength can generally be divided into two main categories (as suggested by Berlow et al. (2004)): 1) the strength of an interaction between individuals of each species, or 2) the effect that changes in one species population has on the dynamics of the other species. It can be measured as the effect over a period of time (in the units of biomass or energy flux (Barnes et al. 2018; Brown et al. 2004)) or the relative importance of one species on another (Heleno et al. 2014; Berlow et al. 2004; Wootton and Emmerson 2005). One recurring observation is that networks are often composed of many weak interactions and few strong interactions (Berlow et al. 2004). The distribution of interaction strength within a network effects its stability (Neutel 2002; Ruiter, Neutel, and Moore 1995) and functioning (Duffy 2002; José M. Montoya, Rodríguez, and Hawkins 2003), and serves to benefit multi-species models (Wootton and Emmerson 2005). Alternatively, understanding flow in modules within networks can aid in understanding the organisation of networks (Farage et al. 2021; Jose M. Montoya and Solé 2002) or the cascading effects of perturbations (Gaiarsa and Guimarães 2019).
In some systems, quantifying interaction strength is relatively straightforward; this includes a lot of host-parasite systems. For example, freshwater cyprinid fish can be divided in micro-habitats (fins, skin, digestive system, gill subsections) and the parasites counted in each of these micro-habitats, giving within-host resolution (Simková et al. 2002); marine sparids and labrids have similarly been studied this way, see notably (Sasal, Niquil, and Bartoli 1999; Desdevises 2006; Morand et al. 2002). In some cases, within-host assessments of interaction strengths can reveal macro-ecological events, like in the conservatism of micro-habitat use in amphibian hosts by helminths (Badets et al. 2011). Even ectoparasites can provide reliable assessments of interaction strength; for example, when rodent hosts are minimally disturbed during capture, fine combing of their fur will result in exhaustive ectoparasites inventories (Hadfield et al. 2014; Karbowiak et al. 2019; Matthee et al. 2020; Sánchez et al. 2014; E. R. Dickinson, Millins, and Biek 2020). Parasites have the desirable property of usually remaining intact within their host during the interaction, as opposed to prey items as can be recovered through e.g. gut content analysis or stable isotopes (Macías-Hernández et al. 2018; Schmid-Araya et al. 2016). As network ecology is starting to explore the use of predictive models, leading up to forecasting, we argue that host-parasite systems can provide data that are reliable and trustworthy enough that they can become the foundations for methodological development and benchmark studies, thereby providing more information about host-parasite systems and supporting the technical development of the field.
Yet in most situations, much like quantifying the occurrence of an interaction, quantifying interaction strength in the field is challenging in the majority of systems, and one must often rely on proxies. In some contexts, interaction strength can be estimated via functional foraging (Portalier et al. 2019), where the primary basis for inferring interaction is foraging behaviour like searching, capture and handling times. In food-webs, metabolic based models use body mass, metabolic demands, and energy loss to infer energy fluxes between organisms (Yodzis and Innes 1992; Berlow et al. 2009). In addition, food-web energetics models can be incorporated at various resolutions for a specific network, ranging from individual-based data to more lumped data at the species level or trophic group, depending on data availability (Barnes et al. 2018; Berlow et al. 2009). Taken together, these considerations impose too many constraints on predicting continuous interaction strength at the moment, resulting in our primary focus in binary present/absent interactions within this manuscript.
How do we determine what interaction networks are feasible?
For several decades, ecologists have aimed to understand how networks of many interacting species persist through time. The diversity-stability paradox, first explored by May (1974), shows that under a neutral set of assumptions ecological networks should become decreasingly stable as the number of species increases. Yet, in the natural world we observe networks of interactions that consist of far more species than May’s model predicts (Albouy et al. 2019). As a result, understanding what aspects of the neutral assumptions of May’s model are incorrect has branched many investigations into the relationship between ecological network structure and persistence (Allesina and Tang 2012). These assumptions can be split into dynamical assumptions and topological assumptions. Topologically, we know that ecological networks are not structured randomly. Some properties, like the aforementioned connectance, are highly predictable (MacDonald, Banville, and Poisot 2020). Generative models of food-webs (based on network embeddings) fit empirical networks more effectively than random models (Allesina, Alonso, and Pascual 2008). These models have long used allometry as a single-dimensional niche space—naturally we want to extend this to traits in general. The second approach to stability is through dynamics. Early models of community dynamics rely on the assumption of linear interaction effects, but in recent years models of bioenergetic community dynamics have shown promise in basing our understanding of energy flow in food-webs in the understood relationship between allometry and metabolism (Delmas et al. 2017). An additional consideration is the multidimensional nature of “stability” and “feasibility” (e.g. resilience to environmental change vs extinctions) (Domínguez-García, Dakos, and Kéfi 2019) and how different disturbances propagate across levels of biological organisation (Kéfi et al. 2019; Gravel, Massol, and Leibold 2016). Recent approaches such as structural stability (Saavedra et al. 2017; Ferrera, Pascual-García, and Bastolla 2016) allow us to think of network feasibility in rigorous mathematical terms, which may end up as usable parameters to penalise network predictions.
What taxonomic scales are suitable for the prediction of species interactions?
If we use different trait-based proxies to predict potential interactions between species the choice of such proxies should be theoretically linked to the taxonomic and spatial scale we are using in our prediction (Wiens 1989). At some scales we can use morphological traits of co-occurring species to assess the probability of interaction between them (Bartomeus et al. 2016). On broader taxonomic scales we can infer interaction probability through the phylogenetic distance, assuming that functional traits themselves are conserved (Gómez, Verdú, and Perfectti 2010). In this case, we can think of the probability that one species will interact with another as the distance between them in niche-space (Desjardins-Proulx et al. 2017), and this can be modelled by simulating neutral expectations of trait variation on phylogenetic trees (Davies 2021). At the narrowest scales, we may be interested in predicting behavioural traits like foraging behaviour (Bartomeus et al. 2016), and at this scale we may need to consider abundance’s effect on the probability of an encounter (Wells and O’Hara 2013).
What about indirect and higher-order interactions?
Although network ecology often assumes that interactions go strictly from one node to the other, the web of life is made up of a variety of interactions. Indirect interactions—either higher-order interactions between species, or interaction strengths that themselves interact — have gained interest in recent years (Golubski et al. 2016; Golubski and Abrams 2011). One mathematical tool to describe these situations is hypergraphs: hypergraphs are the generalisation of a graph, allowing a broad yet manageable approach to complex interactions (Carletti, Fanelli, and Nicoletti 2020), by allowing for particular interactions to occur beyond a pair of nodes. An additional degree of complexity is introduced by multi-layer networks (Hutchinson et al. 2019). Multi-layer networks include edges across “variants” of the networks (timepoints, locations, or environments). These can be particularly useful to account for the metacommunity structure (Gross et al. 2020), or to understand how dispersal can inform conservation action (Albert et al. 2017). Ecological networks are intrinsically multi-layered (Pilosof et al. 2017). However, prima facie, increasing the dimensionality of the object we need to predict (the multiple layers rather than a single network) makes the problem more complicated. Yet, multi-layer approaches improve prediction in social networks (Jalili et al. 2017; Najari et al. 2019; Yasami and Safaei 2018), and they may prove useful in network ecology going forward.
Space
Although networks were initially used to describe the interactions within a community, interest in the last decade has shifted towards understanding their structure and variation over space (Trøjelsgaard and Olesen 2016; Baiser et al. 2019), and has established network ecology as an important emerging component of biogeography and macroecology.
How much do networks vary over space?
Networks can vary across space either in their structural properties (e.g. connectance or degree distribution) or in their composition (identity of nodes and edges). Interestingly, variation in the structural properties of ecological networks primarily responds to changes in the size of the network. The number of links in ecological networks scales with the number of species (MacDonald, Banville, and Poisot 2020; Brose et al. 2004), and connectance and size drive the rest of network structure (Poisot and Gravel 2014; J. A. Dunne, Williams, and Martinez 2002; Riede et al. 2010). Species turnover in space results in changes in the composition of ecological networks. But, this is not the only reason network composition varies (Poisot, Stouffer, and Gravel 2015). Intraspecific variation can result in interaction turnovers without changes in species composition (Bolnick et al. 2011). Similarly, changes in species abundances can lead to variation in interaction strengths (Canard et al. 2014; Vázquez et al. 2007). Variation in the abiotic environment and indirect interactions (Golubski et al. 2016) could modify the occurrence and strength of individual interactions. Despite this, empirical networks tend to share a common backbone (Mora et al. 2018) and functional composition (Dehling et al. 2020) across space.
How do we predict what the species pool at a particular location is?
As the species pool forms the basis for network structure, predicting which species are present at a particular location is essential to predict networks across space. Species distribution models (SDMs) are increasingly ubiquitous in macroecology— these models predict the range of a species based on known occurrences and environmental conditions, such as climate and land cover (Guisan and Thuiller 2005; Elith et al. 2006). Including interactions or co-occurrences in SDMs generally improves predictive performance (Wisz et al. 2013). Several approaches exist to combine multiple SDMs: community assemblage at a particular site can be predicted either by combining independent single-species SDMs (stacked-SDMs, SSDMs) or by directly modelling the entire species assemblage and multiple species at the same time (joint SDMs, JSDMs) (Norberg et al. 2019). Building on the JSDM framework, hierarchical modelling of species communities (Ovaskainen et al. 2017) has the advantage of capturing processes that structure communities. Spatially Explicit Species Assemblage Modelling (SESAM) constrains SDM predictions using macro-ecological models (Guisan and Rahbek 2011) — for example, variation in species richness across space can constrain assemblage predictions (D’Amen et al. 2015).
The next step is to constrain distribution predictions using network properties. This builds on previous calls to adopt a probabilistic view: a probabilistic species pool (Karger et al. 2016), and probabilistic interactions through Bayesian networks (Staniczenko et al. 2017). Blanchet, Cazelles, and Gravel (2020) argue that the probabilistic view avoids confusion between interactions and co-occurrences, but that it requires prior knowledge of interactions. This could potentially be solved through our framework of predicting networks first, interactions next, and finally the realised species pool.
How do we combine spatial and network predictions?
In order to predict networks across space, we need to combine multiple models—one which predicts what the species pool will be at a given location, and one to predict what interaction networks composed from this species pool are likely to be (see fig. 2). Both of these models contain uncertainty, and when we combine them the uncertainty from each model should be propagated into the combined model. The Bayesian paradigm provides a convenient solution to this—if we have a chain of models where each model feeds into the next, we can sample from the posterior of the input models. A different approach is ensemble modelling which combines the predictions made by several models, where each model is predicting the same thing (Parker 2013). Error propagation, an important step in building any ecological model, describes the effect of the uncertainty of input variables on the uncertainty of output variables (Draper 1995; Parysow, Gertner, and Westervelt 2000). Benke et al. (2018) identifies two broad approaches to model error propagation: analytically using differential equations or stochastically using Monte-Carlo simulation methods. Errors induced by the spatial or temporal extrapolation of data also need to be taken into account when estimating the uncertainty of a model’s output (Peters and Herrick 2004).
Time
Why should we forecast species interaction networks?
Forecasting species interactions are critical for informing ecosystem management (Harvey et al. 2017) and systematic conservation prioritisation (Pollock et al. 2020), and for anticipating extinctions and their consequences (McDonald-Madden et al. 2016; McWilliams et al. 2019). Ecological interactions shape species distributions at both local and broad spatial scales, and including interactions in SDM models typically improves predictive performance (M. B. Araújo and Luoto 2007; Wisz et al. 2013; Pigot and Tobias 2013). However, these tend to rely on approaches involving estimating pairwise dependencies based on co-occurrence, using surrogates for biotic-interaction gradients, and hybridising SDMs with dynamic models (Wisz et al. 2013). Most existing models to predict the future distribution of species ignore interactions (Urban et al. 2016). Changes in species ranges and phenology will inevitably create spatiotemporal mismatches and affect encounter rates between species (Gilman et al. 2010), which will further shift the distribution of species across space. New interactions will also appear between species that are not currently co-occurring (Gilman et al. 2010). Only by forecasting how species will interact can we hope to have an accurate portrait of how biodiversity will be distributed under the future climate.
Forecasting how climate change will alter biodiversity is also crucial for maximising conservation outcomes. Improving SDMs through interactions is crucial for conservation, as nearly 30% of models in SDM studies are used to assess population declines or landscape ability to support populations (M. B. Araújo et al. 2019). Reliable predictions about how ecological networks will change over time will give us critical information that could be communicated to decision-makers and the scientific community about what future environmental risks we are awaiting and how to mitigate them (Kindsvater et al. 2018). Not only this, but how biodiversity is structured influences the functioning of the whole ecosystem, community stability and persistence (Thompson et al. 2012; Stouffer and Bascompte 2010). Will climate change impact the distribution of network properties (e.g. connectance)? If so, which regions or species groups need special conservation efforts? These overarching questions are yet to be answered (but see Albouy et al. 2013; Kortsch et al. 2015; Hattab et al. 2016). We believe that the path toward forecasting ecological networks provides useful guidelines to ultimately better predict how climate change will affect the different dimensions of biodiversity and ecosystem functioning.
How do we turn a predictive model into a forecasting model?
On some scales, empirical time-series encode enough information about ecological processes for machine-learning approaches to make accurate forecasts. However, there is an intrinsic limit to the predictability of ecological time-series (Pennekamp et al. 2019). A forecast inherently has a resolution limit in space, time, and organisation. For example, one could never hope to predict the precise abundance of every species on Earth on every day hundreds of years into the future. There is often a trade-off between the resolution and horizon of forecast, e.g., a lower resolution forecast, like primary production will be at a maximum in the summer, is likely to be true much further into the future than a higher resolution forecast. If we want to forecast the structure of ecological networks beyond the forecasting horizon of time-series based methods, we need forecasts of our predictive model’s inputs—a forecast of the distribution of both environmental conditions and the potential species pool across space (fig. 3).
How can we validate a forecasting model?
Often the purpose of building a forecasting model is to inform present action (Dietze et al. 2018). Yet, the nature of forecasting—trying to predict the future—is that you can only know if a forecast is “right” once it is too late to change it. If we want to maximise the chance that reality falls within a forecasting model’s predictions, there are two directions to approach this problem: the first is to extend model validation techniques to a forecasting context, and the second is to attempt to maximise the amount of uncertainty in the forecast without compromising its resolution. Cross-validation (see How do we validate a predictive model?) can be used to test the efficacy of a forecasting model. Given a time-series of N observations, a model can iteratively be trained on the first n time-points of data, and the forecasting model’s accuracy can be evaluated on the remaining time-points it hasn’t “seen” (Bishop 2006). This enables us to understand both how much temporal data is required for a model to be robust, and also enables us to explore the forecasting horizon of a process. Further, this approach can also be applied in the opposite temporal direction— if we have reliable data from the past, “hindcasting” can also be used to test a forecast’s robustness.
However, these methods inevitably bump into a hard-limitation on what is feasible for a forecasting model. The future is uncertain. Any empirical time-series we use to validate a model was collected in past conditions that may not persist into the future. Any system we wish to forecast will undergo only one of many possible scenarios, yet we can only observe the realised outcome of the system under the scenario that actually unfolds. It is therefore impossible to assess the quality of a forecasting model in scenarios that remain hypothetical. If the goal is to maximise the probability that reality will fall within the forecast’s estimates, forecasts should incorporate as much uncertainty about the future scenario as possible—one way to do this is ensemble modelling (Parker 2013). However, as we increase the amount of uncertainty we incorporate into a forecasting model, the resolution of the forecast’s predictions could shrink (Lei and Whitaker 2017), and therefore the modeller should be mindful of the trade-off between resolution and accuracy when developing any forecast. Finally, ensemble models are not guaranteed to give more accurate results: for example, Becker et al. (2020) noted that the ensemble model outperforms the best-in-class models, which should be taken as an indication that careful model building and selection is of the utmost importance when dealing with a problem as complex as the prediction of species interactions.
Conclusion: why should we predict species interaction networks?
Because we almost can, and because we definitely should.
A better understanding of species interactions, and the networks they form, would help unify the fields of community, network, and spatial ecology; improve the quantification of the functional relationships between species (Dehling and Stouffer 2018; O’Connor et al. 2020); re-evaluate metacommunities in light of network structure (Guzman et al. 2019); and enable a new line of research into the biogeography of species interactions (Massol et al. 2017; Braga et al. 2019) which incorporates a synthesis of both Eltonian and Grinnellian niche (Gravel et al. 2019). Further, the ability to reliably predict and forecast species interactions would inform conservation efforts for protecting species, communities, and ecosystems. Integration of species interactions into the assessment of vulnerability to climate change is a needed methodological advancement (Foden and Young 2016). International panels draw on models to establish scientific consensus (M. B. Araújo et al. 2019), and they can be improved through more effective prediction of species distributions and interactions (Syfert et al. 2014). Further, recent studies argue for a shift in focus from species to interaction networks for biodiversity conservation to better understand ecosystem processes (Harvey et al. 2017).
We should invest in network prediction because the right conditions to do so reliably and rapidly are beginning to emerge. Given the possible benefits to a variety of ecological disciplines that would result from an increased ability to predict networks, we feel strongly that the research agenda we outline here should be picked up by the community. Although novel technologies are bringing massive amounts of data to some parts of ecology (primarily environmental DNA and remote sensing, but now more commonly image analysis and bioacoustics), it is even more important to be intentional about reconciling data. This involves not only the work of understanding the processes encoded within data, but also the groundwork of developing pipelines to bridge the ever-expanding gap between “high-throughput” and “low-throughput” sampling methods. An overall increase in the volume of data will not result in an increase of our predictive capacity as long as this data increase is limited to specific aspects of the problem. In the areas we highlight in fig. 2, many data steps are still limiting: documenting empirical interactions is natural history work that doesn’t lend itself to systematic automation; expert knowledge is by design a social process that may be slightly accelerated by text mining and natural language processing (but is not yet, or not routinely or at scale). These limitations are affecting our ability to reconstruct networks.
But the tools to which we feed these data, incomplete as they may be, are gradually getting better; that is, they can do predictions faster, they handle uncertainty and propagate it well, and they can accommodate data volumes that are lower than we may expect (Pichler et al. 2020). It is clear attempting to predict the structure of ecological networks at any scale is a methodological and ecological challenge; yet it will result in qualitative changes in our understanding of complex adaptive systems, as well as changes to our ability to leverage information about network structure for conservation decision. It is perhaps even more important to forecast the structure of ecological networks because it is commonly neglected as a facet of biodiversity that can (and should) be managed. In fact, none of the Aichi targets mention biostructure or its protection, despite this being recognised as an important task (McCann 2007), either implicitly or explicitly. Being able to generate reliable datasets on networks in space or time will make this information more actionable.
Acknowledgements: We acknowledge that this study was conducted on land within the traditional unceded territory of the Saint Lawrence Iroquoian, Anishinabewaki, Mohawk, Huron-Wendat, and Omàmiwininiwak nations. TS, NF, TP are funded by a donation from the Courtois Foundation; FB, NF, and TP are funded by IVADO; BM is funded by the NSERC Alexander Graham Bell Canada Graduate Scholarship and the FRQNT master’s scholarship; FB, GD, NF, and GH are funded by the NSERC BIOS2 CREATE program; GD is funded by the FRQNT doctoral scholarship; DC, TS, LP, and TP are funded by the Canadian Institute of Ecology & Evolution; this research was enabled in part by support provided by Calcul Québec (www.calculquebec.ca) and Compute Canada (www.computecanada.ca). This work was supported by funding to the Viral Emergence Research Initiative (VERENA) consortium including NSF BII 2021909. AG and MDC are supported in part by the Liber Ero Chair.