Zonal statistics
In this tutorial, we will grab some bioclimatic variables for New Zealand, and then identify the districts that have extreme values of this variable.
using SpeciesDistributionToolkit
using Statistics
using CairoMakieWe will get the BIO19 layer from CHELSA2 (most precipitation in the coldest quarter):
spatial_extent =
(left = 165.739746, bottom = -47.587547, right = 180.812988, top = -33.649514)
dataprovider = RasterData(CHELSA2, BioClim)
layer = SDMLayer(dataprovider; layer = "BIO19", spatial_extent...)SDM Layer with 2865888 UInt16 cells
Proj string: +proj=longlat +datum=WGS84 +no_defs
Grid size: (1674, 1712)This layer is trimmed to the landmass (according to GADM):
mask!(layer, SpeciesDistributionToolkit.gadm("NZL"))
layer = trim(layer)SDM Layer with 417341 UInt16 cells
Proj string: +proj=longlat +datum=WGS84 +no_defs
Grid size: (1578, 1455)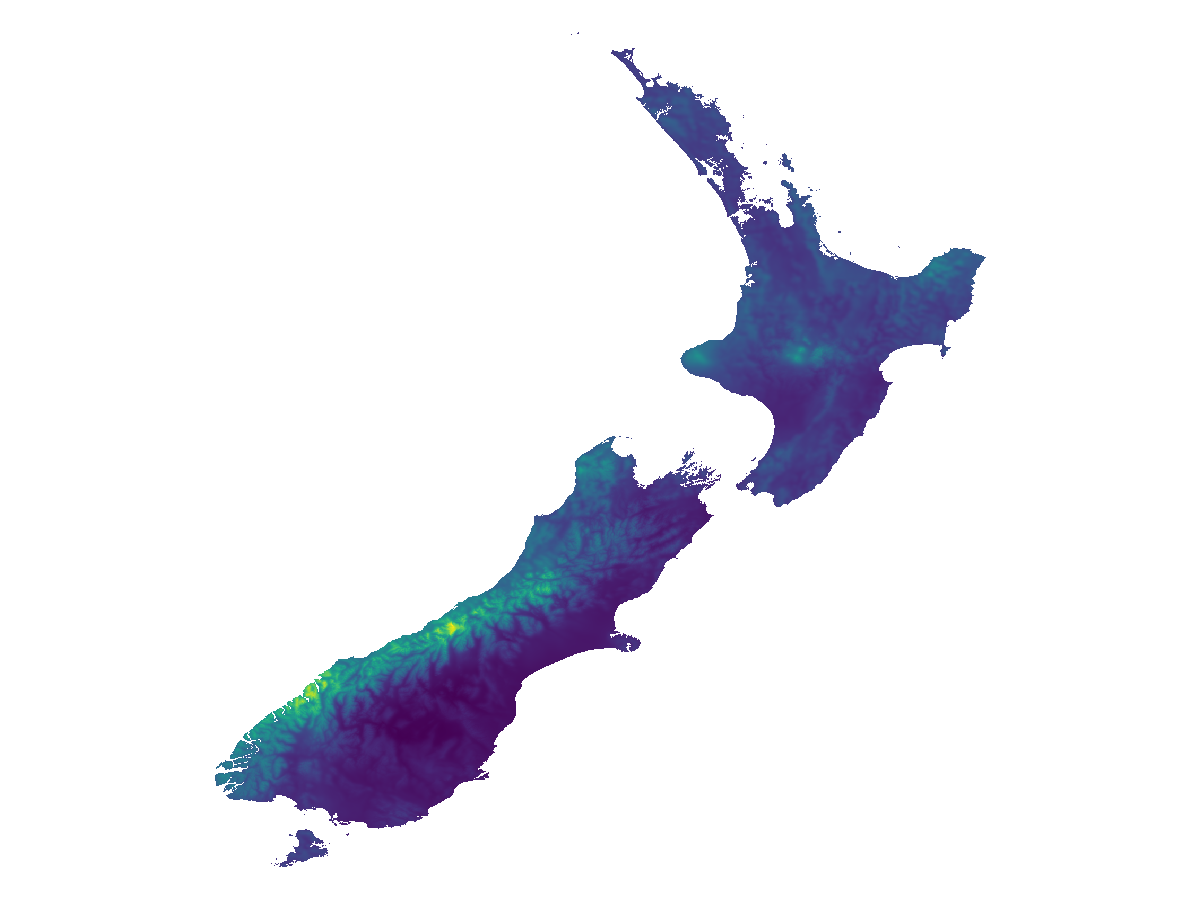
Code for the figure
fig, ax, plt = heatmap(layer; axis = (; aspect = DataAspect()))
hidespines!(ax)
hidedecorations!(ax)We can now get the lower level sub-division. Note that not all territories covered by GADM have the same number of sub-divisions!
districts = SpeciesDistributionToolkit.gadm("NZL", 2);We can start looking at how these map onto the landscape, using the zone function. It will return a layer where the value of each pixel is the index of the polygon containing this pixel:
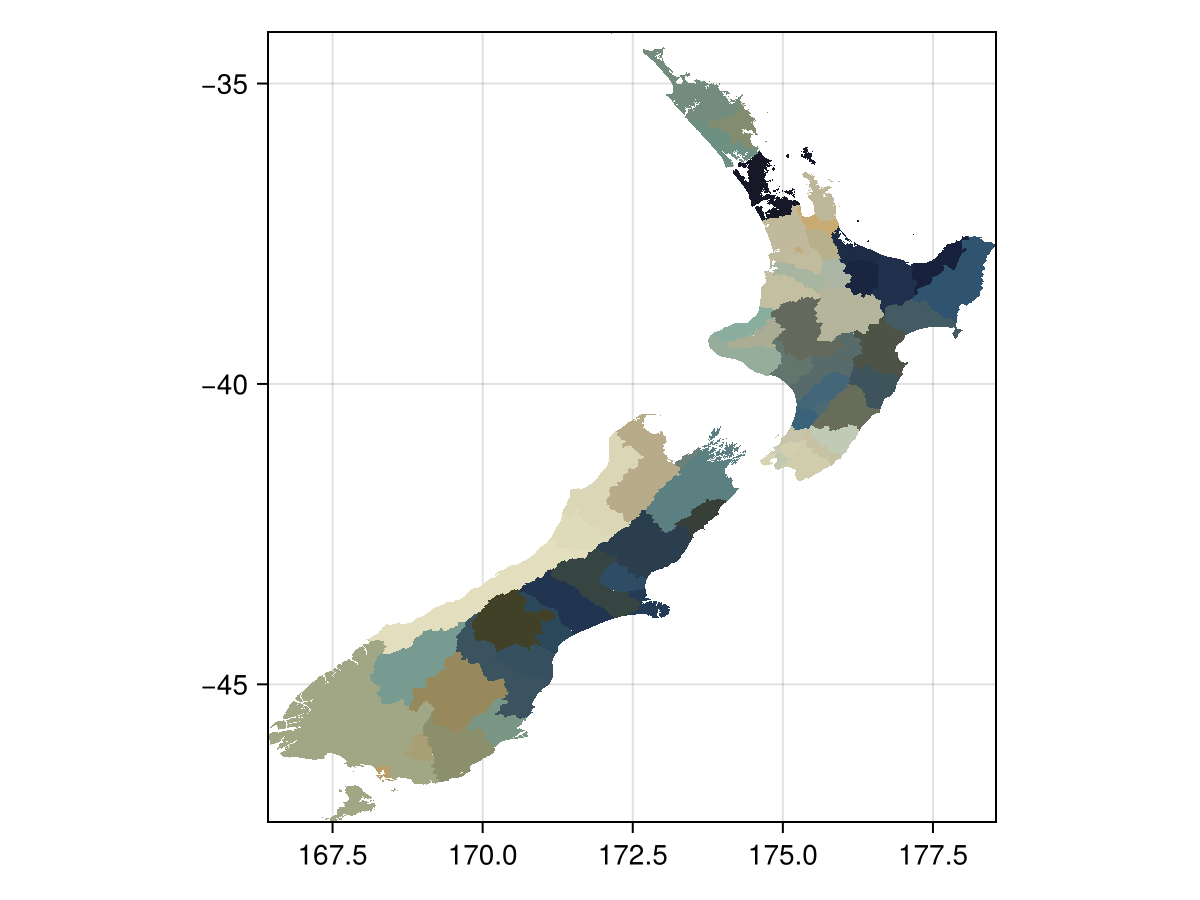
Code for the figure
heatmap(zone(layer, districts); colormap = :hokusai, axis = (; aspect = DataAspect()))Note that the pixels that are not within a polygon are turned off, which can sometimes happen if the overlap between polygons is not perfect. There is a variant of the mosaic method that uses polygon to assign the values:
z = mosaic(median, layer, districts)
nodata!(z, 0.0)SDM Layer with 417284 Float64 cells
Proj string: +proj=longlat +datum=WGS84 +no_defs
Grid size: (1578, 1455)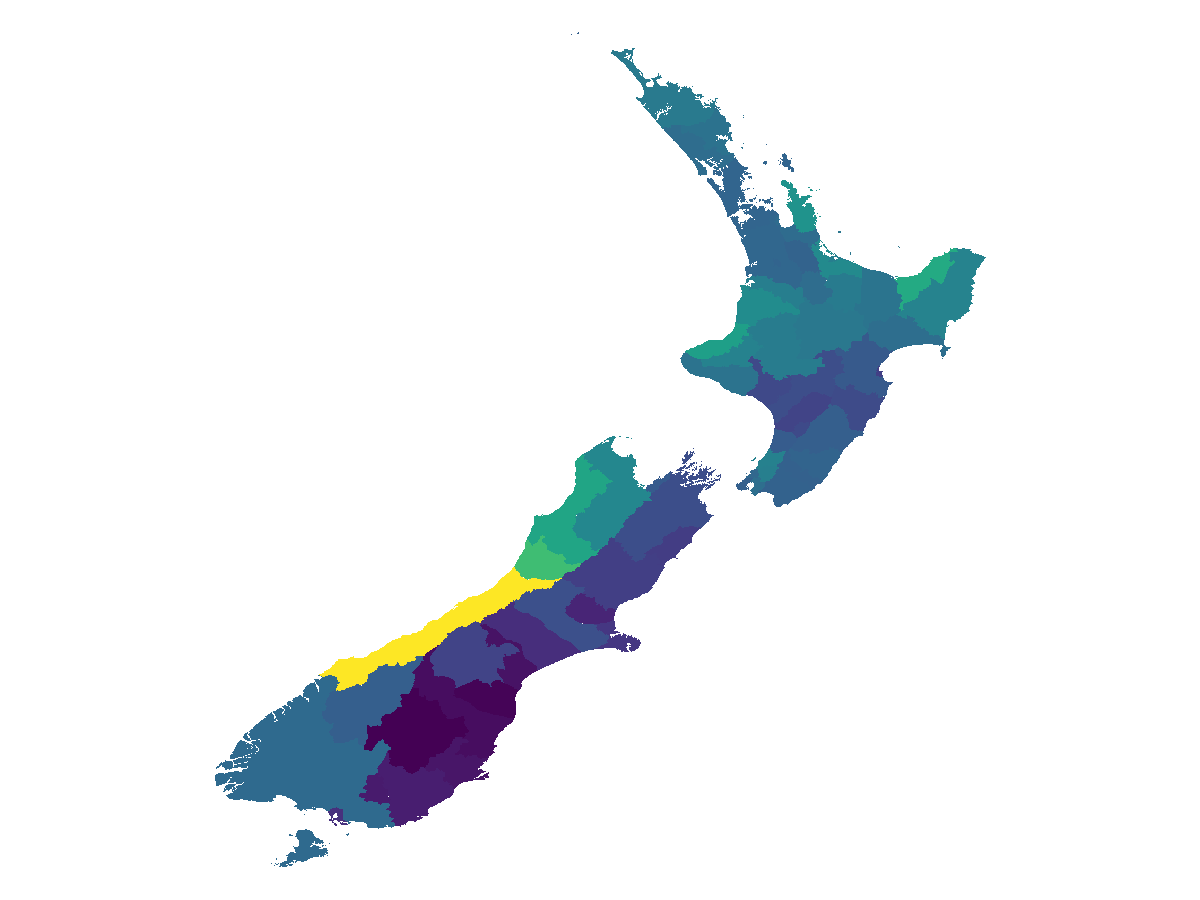
Code for the figure
fig, ax, plt = heatmap(z; axis = (; aspect = DataAspect()))
hidespines!(ax)
hidedecorations!(ax)In order to make a plot identifying some areas, we get their full names using gadmlist:
districtnames = SpeciesDistributionToolkit.gadmlist("NZL", 2)
districtnames[1:10]10-element Vector{String}:
"Auckland"
"AreaOutsideTerritorialAuthori"
"Kawerau"
"Opotiki"
"Rotorua"
"Tauranga"
"WesternBayofPlenty"
"Whakatane"
"Ashburton"
"Christchurch"Finally, we can get the median value within each of these polygons using the byzone method:
top5 =
first.(
sort(
byzone(median, layer, districts, districtnames);
by = (x) -> x.second,
rev = true,
)[1:5]
)5-element Vector{String}:
"Westland"
"Grey"
"Opotiki"
"Buller"
"NewPlymouth"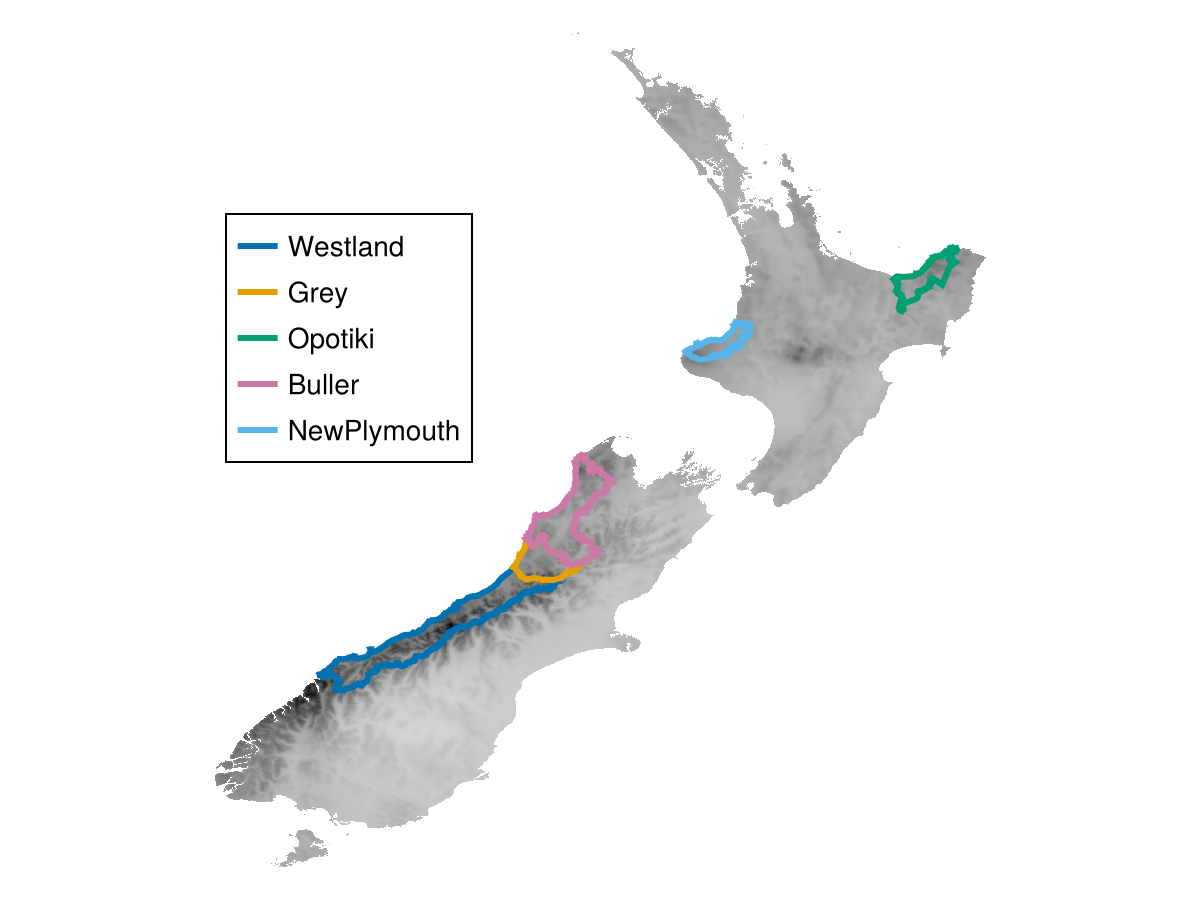
Code for the figure
fig, ax, plt =
heatmap(layer; axis = (; aspect = DataAspect()), colormap = [:lightgrey, :black])
[
lines!(ax, districts[i]; label = districtnames[i], linewidth = 3) for
i in indexin(top5, districtnames)
]
axislegend(; position = (0, 0.7), nbanks = 1)